Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
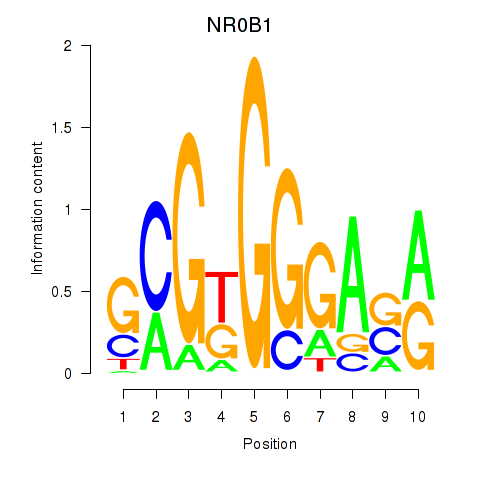
Results for NR0B1
Z-value: 0.91
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | NR0B1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR0B1 | hg19_v2_chrX_-_30326445_30326605, hg19_v2_chrX_-_30327495_30327509 | -0.24 | 3.8e-01 | Click! |
Activity profile of NR0B1 motif
Sorted Z-values of NR0B1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR0B1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_56338750 | 1.87 |
ENST00000345724.3 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr9_+_128509624 | 1.80 |
ENST00000342287.5 ENST00000373487.4 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
| chr6_-_29595779 | 1.64 |
ENST00000355973.3 ENST00000377012.4 |
GABBR1 |
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr2_-_60780702 | 1.61 |
ENST00000359629.5 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr2_-_60780607 | 1.49 |
ENST00000537768.1 ENST00000335712.6 ENST00000356842.4 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr6_+_7541808 | 1.37 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr6_+_7541845 | 1.36 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr19_-_45909585 | 1.21 |
ENST00000593226.1 ENST00000418234.2 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
| chrX_-_154688276 | 1.16 |
ENST00000369445.2 |
F8A3 |
coagulation factor VIII-associated 3 |
| chr19_-_2050852 | 1.08 |
ENST00000541165.1 ENST00000591601.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr19_+_8429031 | 1.02 |
ENST00000301455.2 ENST00000541807.1 ENST00000393962.2 |
ANGPTL4 |
angiopoietin-like 4 |
| chr2_-_60780536 | 1.01 |
ENST00000538214.1 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
| chr1_-_93426998 | 0.96 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr9_+_128509663 | 0.96 |
ENST00000373489.5 ENST00000373483.2 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
| chr6_+_391739 | 0.96 |
ENST00000380956.4 |
IRF4 |
interferon regulatory factor 4 |
| chr7_+_69064300 | 0.95 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr8_+_26435359 | 0.92 |
ENST00000311151.5 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr19_-_2051223 | 0.85 |
ENST00000309340.7 ENST00000589534.1 ENST00000250896.3 ENST00000589509.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr13_-_52027134 | 0.84 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr3_-_129612394 | 0.83 |
ENST00000505616.1 ENST00000426664.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr2_+_234545092 | 0.82 |
ENST00000344644.5 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr8_+_95653302 | 0.81 |
ENST00000423620.2 ENST00000433389.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr6_+_116692102 | 0.74 |
ENST00000359564.2 |
DSE |
dermatan sulfate epimerase |
| chr12_+_56473628 | 0.72 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr11_-_119993979 | 0.70 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_2160180 | 0.67 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr20_+_46130601 | 0.65 |
ENST00000341724.6 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr12_+_56473939 | 0.63 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr2_+_54198210 | 0.57 |
ENST00000607452.1 ENST00000422521.2 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr2_+_201171064 | 0.56 |
ENST00000451764.2 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr1_-_147245484 | 0.56 |
ENST00000271348.2 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr12_-_6484715 | 0.56 |
ENST00000228916.2 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr17_+_76165213 | 0.55 |
ENST00000590201.1 |
SYNGR2 |
synaptogyrin 2 |
| chr3_-_11623804 | 0.54 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chrX_-_130423386 | 0.53 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr3_+_133292851 | 0.52 |
ENST00000503932.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr20_+_55204351 | 0.51 |
ENST00000201031.2 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr8_+_95653427 | 0.50 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr1_-_154600421 | 0.49 |
ENST00000368471.3 ENST00000292205.5 |
ADAR |
adenosine deaminase, RNA-specific |
| chr2_+_54342574 | 0.47 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr10_+_112257596 | 0.46 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr2_-_152955537 | 0.46 |
ENST00000201943.5 ENST00000539935.1 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr17_+_7211280 | 0.46 |
ENST00000419711.2 ENST00000571955.1 ENST00000573714.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr5_+_67511524 | 0.46 |
ENST00000521381.1 ENST00000521657.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_+_44401479 | 0.45 |
ENST00000438616.3 |
ARTN |
artemin |
| chr13_+_25670268 | 0.43 |
ENST00000281589.3 |
PABPC3 |
poly(A) binding protein, cytoplasmic 3 |
| chr15_+_74833518 | 0.43 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chrX_-_130423240 | 0.42 |
ENST00000370910.1 ENST00000370901.4 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr1_-_19229014 | 0.42 |
ENST00000538839.1 ENST00000290597.5 |
ALDH4A1 |
aldehyde dehydrogenase 4 family, member A1 |
| chr15_-_83953466 | 0.41 |
ENST00000345382.2 |
BNC1 |
basonuclin 1 |
| chr9_+_137218362 | 0.40 |
ENST00000481739.1 |
RXRA |
retinoid X receptor, alpha |
| chr8_+_61591337 | 0.40 |
ENST00000423902.2 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
| chr20_-_18038521 | 0.39 |
ENST00000278780.6 |
OVOL2 |
ovo-like zinc finger 2 |
| chr2_+_234580499 | 0.39 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr1_-_46598371 | 0.38 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr17_-_7155274 | 0.37 |
ENST00000318988.6 ENST00000575783.1 ENST00000573600.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr1_-_156390128 | 0.36 |
ENST00000368242.3 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr18_-_51750948 | 0.36 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr12_+_56473910 | 0.36 |
ENST00000411731.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr2_+_234580525 | 0.36 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr17_-_40761375 | 0.34 |
ENST00000543197.1 ENST00000309428.5 |
FAM134C |
family with sequence similarity 134, member C |
| chr17_+_78075361 | 0.33 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr2_-_152955213 | 0.33 |
ENST00000427385.1 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr5_+_110559603 | 0.33 |
ENST00000512453.1 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr17_-_7155802 | 0.32 |
ENST00000572043.1 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
| chr4_-_151936865 | 0.32 |
ENST00000535741.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chrX_-_153151586 | 0.31 |
ENST00000370060.1 ENST00000370055.1 ENST00000420165.1 |
L1CAM |
L1 cell adhesion molecule |
| chr12_+_103351444 | 0.31 |
ENST00000266744.3 |
ASCL1 |
achaete-scute family bHLH transcription factor 1 |
| chr1_+_180601139 | 0.31 |
ENST00000367590.4 ENST00000367589.3 |
XPR1 |
xenotropic and polytropic retrovirus receptor 1 |
| chr5_+_175223715 | 0.30 |
ENST00000515502.1 |
CPLX2 |
complexin 2 |
| chr2_+_234545148 | 0.30 |
ENST00000373445.1 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_+_219081817 | 0.30 |
ENST00000315717.5 ENST00000420104.1 ENST00000295685.10 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr2_+_232573208 | 0.29 |
ENST00000409115.3 |
PTMA |
prothymosin, alpha |
| chr14_-_21566731 | 0.29 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr2_+_54342533 | 0.29 |
ENST00000406041.1 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr3_-_13921594 | 0.29 |
ENST00000285018.4 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
| chr10_+_38299546 | 0.28 |
ENST00000374618.3 ENST00000432900.2 ENST00000458705.2 ENST00000469037.2 |
ZNF33A |
zinc finger protein 33A |
| chr9_+_71320596 | 0.28 |
ENST00000265382.3 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr20_+_57466357 | 0.28 |
ENST00000371095.3 ENST00000371085.3 ENST00000354359.7 ENST00000265620.7 |
GNAS |
GNAS complex locus |
| chr19_-_49576198 | 0.28 |
ENST00000221444.1 |
KCNA7 |
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr17_-_2614927 | 0.28 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr15_+_74422585 | 0.27 |
ENST00000561740.1 ENST00000435464.1 ENST00000453268.2 |
ISLR2 |
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr17_-_4890919 | 0.27 |
ENST00000572543.1 ENST00000381311.5 ENST00000348066.3 ENST00000358183.4 |
CAMTA2 |
calmodulin binding transcription activator 2 |
| chr20_+_57466629 | 0.27 |
ENST00000371081.1 ENST00000338783.6 |
GNAS |
GNAS complex locus |
| chr17_+_29421987 | 0.27 |
ENST00000431387.4 |
NF1 |
neurofibromin 1 |
| chr20_+_43104508 | 0.27 |
ENST00000262605.4 ENST00000372904.3 |
TTPAL |
tocopherol (alpha) transfer protein-like |
| chr4_+_38665810 | 0.27 |
ENST00000261438.5 ENST00000514033.1 |
KLF3 |
Kruppel-like factor 3 (basic) |
| chr11_+_76092353 | 0.26 |
ENST00000530460.1 ENST00000321844.4 |
RP11-111M22.2 |
Homo sapiens putative uncharacterized protein FLJ37770-like (LOC100506127), mRNA. |
| chr20_+_35201857 | 0.25 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr2_+_204193149 | 0.25 |
ENST00000422511.2 |
ABI2 |
abl-interactor 2 |
| chr4_-_151936416 | 0.25 |
ENST00000510413.1 ENST00000507224.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr5_-_180237445 | 0.25 |
ENST00000393340.3 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr3_+_171758344 | 0.25 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr11_+_64863587 | 0.25 |
ENST00000530773.1 ENST00000279281.3 ENST00000529180.1 |
VPS51 |
vacuolar protein sorting 51 homolog (S. cerevisiae) |
| chr2_-_44223138 | 0.25 |
ENST00000260665.7 |
LRPPRC |
leucine-rich pentatricopeptide repeat containing |
| chr1_-_236030216 | 0.24 |
ENST00000389794.3 ENST00000389793.2 |
LYST |
lysosomal trafficking regulator |
| chr6_+_64281906 | 0.24 |
ENST00000370651.3 |
PTP4A1 |
protein tyrosine phosphatase type IVA, member 1 |
| chr2_-_74405929 | 0.24 |
ENST00000396049.4 |
MOB1A |
MOB kinase activator 1A |
| chrX_-_112084043 | 0.24 |
ENST00000304758.1 |
AMOT |
angiomotin |
| chr5_-_142783694 | 0.24 |
ENST00000394466.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr6_+_37787262 | 0.23 |
ENST00000287218.4 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
| chr16_-_28937027 | 0.23 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr17_-_4269768 | 0.23 |
ENST00000396981.2 |
UBE2G1 |
ubiquitin-conjugating enzyme E2G 1 |
| chr17_+_40811283 | 0.23 |
ENST00000251412.7 |
TUBG2 |
tubulin, gamma 2 |
| chr8_-_125486755 | 0.23 |
ENST00000499418.2 ENST00000530778.1 |
RNF139-AS1 |
RNF139 antisense RNA 1 (head to head) |
| chr2_+_85132749 | 0.23 |
ENST00000233143.4 |
TMSB10 |
thymosin beta 10 |
| chr9_+_79792269 | 0.23 |
ENST00000376634.4 ENST00000376636.3 ENST00000360280.3 |
VPS13A |
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr17_+_4736627 | 0.23 |
ENST00000355280.6 ENST00000347992.7 |
MINK1 |
misshapen-like kinase 1 |
| chr17_+_46018872 | 0.22 |
ENST00000583599.1 ENST00000434554.2 ENST00000225573.4 ENST00000544840.1 ENST00000534893.1 |
PNPO |
pyridoxamine 5'-phosphate oxidase |
| chr3_-_100120223 | 0.22 |
ENST00000284320.5 |
TOMM70A |
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr1_+_39456895 | 0.22 |
ENST00000432648.3 ENST00000446189.2 ENST00000372984.4 |
AKIRIN1 |
akirin 1 |
| chr1_-_94312706 | 0.22 |
ENST00000370244.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chrX_+_133507327 | 0.22 |
ENST00000332070.3 ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6 |
PHD finger protein 6 |
| chr21_-_32931290 | 0.22 |
ENST00000286827.3 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
| chr1_-_151431909 | 0.21 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr3_+_73045936 | 0.21 |
ENST00000356692.5 ENST00000488810.1 ENST00000394284.3 ENST00000295862.9 ENST00000495566.1 |
PPP4R2 |
protein phosphatase 4, regulatory subunit 2 |
| chr2_+_204193129 | 0.21 |
ENST00000417864.1 |
ABI2 |
abl-interactor 2 |
| chr22_-_38794490 | 0.21 |
ENST00000400206.2 |
CSNK1E |
casein kinase 1, epsilon |
| chr6_+_43149903 | 0.21 |
ENST00000252050.4 ENST00000354495.3 ENST00000372647.2 |
CUL9 |
cullin 9 |
| chr7_+_94139105 | 0.20 |
ENST00000297273.4 |
CASD1 |
CAS1 domain containing 1 |
| chr14_+_57046500 | 0.20 |
ENST00000261556.6 |
TMEM260 |
transmembrane protein 260 |
| chr1_-_109584716 | 0.20 |
ENST00000531337.1 ENST00000529074.1 ENST00000369965.4 |
WDR47 |
WD repeat domain 47 |
| chr1_-_46598284 | 0.20 |
ENST00000423209.1 ENST00000262741.5 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr1_+_211433275 | 0.20 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr14_+_57046530 | 0.19 |
ENST00000536419.1 ENST00000538838.1 |
TMEM260 |
transmembrane protein 260 |
| chr17_+_46184911 | 0.19 |
ENST00000580219.1 ENST00000452859.2 ENST00000393405.2 ENST00000439357.2 ENST00000359238.2 |
SNX11 |
sorting nexin 11 |
| chrX_-_119445306 | 0.19 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr4_+_83351791 | 0.19 |
ENST00000509635.1 |
ENOPH1 |
enolase-phosphatase 1 |
| chr6_+_7107999 | 0.19 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr11_-_18343669 | 0.18 |
ENST00000396253.3 ENST00000349215.3 ENST00000438420.2 |
HPS5 |
Hermansky-Pudlak syndrome 5 |
| chr4_+_17616253 | 0.18 |
ENST00000237380.7 |
MED28 |
mediator complex subunit 28 |
| chr20_+_306177 | 0.18 |
ENST00000544632.1 |
SOX12 |
SRY (sex determining region Y)-box 12 |
| chr17_-_77770830 | 0.18 |
ENST00000269385.4 |
CBX8 |
chromobox homolog 8 |
| chr2_+_191513789 | 0.18 |
ENST00000409581.1 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr1_-_40098672 | 0.18 |
ENST00000535435.1 |
HEYL |
hes-related family bHLH transcription factor with YRPW motif-like |
| chr4_-_2758015 | 0.17 |
ENST00000510267.1 ENST00000503235.1 ENST00000315423.7 |
TNIP2 |
TNFAIP3 interacting protein 2 |
| chr15_-_66858298 | 0.17 |
ENST00000537670.1 |
LCTL |
lactase-like |
| chr2_+_97203082 | 0.17 |
ENST00000454558.2 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr17_-_65241281 | 0.17 |
ENST00000358691.5 ENST00000580168.1 |
HELZ |
helicase with zinc finger |
| chr12_+_41086297 | 0.17 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr1_-_40041925 | 0.17 |
ENST00000372862.3 |
PABPC4 |
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr16_-_10674528 | 0.16 |
ENST00000359543.3 |
EMP2 |
epithelial membrane protein 2 |
| chr2_+_217277137 | 0.16 |
ENST00000430374.1 ENST00000357276.4 ENST00000444508.1 |
SMARCAL1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr14_-_21905395 | 0.16 |
ENST00000430710.3 ENST00000553283.1 |
CHD8 |
chromodomain helicase DNA binding protein 8 |
| chr2_-_43823093 | 0.16 |
ENST00000405006.4 |
THADA |
thyroid adenoma associated |
| chr17_-_4890649 | 0.16 |
ENST00000361571.5 |
CAMTA2 |
calmodulin binding transcription activator 2 |
| chr2_-_43823119 | 0.16 |
ENST00000403856.1 ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA |
thyroid adenoma associated |
| chr19_+_1205740 | 0.15 |
ENST00000326873.7 |
STK11 |
serine/threonine kinase 11 |
| chr12_-_54020107 | 0.15 |
ENST00000588232.1 ENST00000548446.2 ENST00000420353.2 ENST00000591397.1 ENST00000415113.1 |
ATF7 |
activating transcription factor 7 |
| chr17_-_42200958 | 0.15 |
ENST00000336057.5 |
HDAC5 |
histone deacetylase 5 |
| chr8_-_144679532 | 0.15 |
ENST00000534380.1 ENST00000533494.1 ENST00000531218.1 ENST00000526340.1 ENST00000533204.1 ENST00000532400.1 ENST00000529516.1 ENST00000534377.1 ENST00000531621.1 ENST00000530191.1 ENST00000524900.1 ENST00000526838.1 ENST00000531931.1 ENST00000534475.1 ENST00000442189.2 ENST00000524624.1 ENST00000532596.1 ENST00000529832.1 ENST00000530306.1 ENST00000530545.1 ENST00000525261.1 ENST00000534804.1 ENST00000528303.1 ENST00000528610.1 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr8_+_128748308 | 0.15 |
ENST00000377970.2 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
| chr19_-_4124079 | 0.15 |
ENST00000394867.4 ENST00000262948.5 |
MAP2K2 |
mitogen-activated protein kinase kinase 2 |
| chr22_+_32870661 | 0.15 |
ENST00000266087.7 |
FBXO7 |
F-box protein 7 |
| chr8_+_28174496 | 0.15 |
ENST00000518479.1 |
PNOC |
prepronociceptin |
| chr17_+_29421900 | 0.14 |
ENST00000358273.4 ENST00000356175.3 |
NF1 |
neurofibromin 1 |
| chr1_+_11751748 | 0.14 |
ENST00000294485.5 |
DRAXIN |
dorsal inhibitory axon guidance protein |
| chr1_+_6052700 | 0.14 |
ENST00000378092.1 ENST00000445501.1 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr3_-_125094093 | 0.13 |
ENST00000484491.1 ENST00000492394.1 ENST00000471196.1 ENST00000468369.1 ENST00000544464.1 ENST00000485866.1 ENST00000360647.4 |
ZNF148 |
zinc finger protein 148 |
| chr6_+_7107830 | 0.13 |
ENST00000379933.3 |
RREB1 |
ras responsive element binding protein 1 |
| chr19_+_36132631 | 0.13 |
ENST00000379026.2 ENST00000379023.4 ENST00000402764.2 ENST00000479824.1 |
ETV2 |
ets variant 2 |
| chr19_+_532049 | 0.13 |
ENST00000606136.1 |
CDC34 |
cell division cycle 34 |
| chr2_-_165698521 | 0.13 |
ENST00000409184.3 ENST00000392717.2 ENST00000456693.1 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr5_+_175298674 | 0.13 |
ENST00000514150.1 |
CPLX2 |
complexin 2 |
| chr2_+_232573222 | 0.13 |
ENST00000341369.7 ENST00000409683.1 |
PTMA |
prothymosin, alpha |
| chr17_+_40761660 | 0.13 |
ENST00000251413.3 ENST00000591509.1 |
TUBG1 |
tubulin, gamma 1 |
| chr10_-_112064665 | 0.13 |
ENST00000369603.5 |
SMNDC1 |
survival motor neuron domain containing 1 |
| chr8_-_101734170 | 0.13 |
ENST00000522387.1 ENST00000518196.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr2_+_204192942 | 0.12 |
ENST00000295851.5 ENST00000261017.5 |
ABI2 |
abl-interactor 2 |
| chr2_+_220492116 | 0.12 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr8_+_22132810 | 0.12 |
ENST00000356766.6 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
| chr7_-_151329416 | 0.12 |
ENST00000418337.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr8_-_102217796 | 0.12 |
ENST00000519744.1 ENST00000311212.4 ENST00000521272.1 ENST00000519882.1 |
ZNF706 |
zinc finger protein 706 |
| chr5_+_175298573 | 0.12 |
ENST00000512824.1 |
CPLX2 |
complexin 2 |
| chr1_+_197170592 | 0.12 |
ENST00000535699.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr5_+_175792459 | 0.12 |
ENST00000310389.5 |
ARL10 |
ADP-ribosylation factor-like 10 |
| chrX_+_9754461 | 0.12 |
ENST00000380913.3 |
SHROOM2 |
shroom family member 2 |
| chr12_+_12764773 | 0.12 |
ENST00000228865.2 |
CREBL2 |
cAMP responsive element binding protein-like 2 |
| chr1_-_41131326 | 0.11 |
ENST00000372684.3 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
| chr1_+_43148625 | 0.11 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
| chr8_+_22132847 | 0.11 |
ENST00000521356.1 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
| chr7_-_155604967 | 0.11 |
ENST00000297261.2 |
SHH |
sonic hedgehog |
| chr2_-_219925189 | 0.11 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr6_+_126070726 | 0.11 |
ENST00000368364.3 |
HEY2 |
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr2_+_220492373 | 0.11 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr2_+_166152283 | 0.11 |
ENST00000375427.2 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr12_+_109535373 | 0.10 |
ENST00000242576.2 |
UNG |
uracil-DNA glycosylase |
| chr3_+_128720424 | 0.10 |
ENST00000480450.1 ENST00000436022.2 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
| chr21_-_15755446 | 0.10 |
ENST00000544452.1 ENST00000285667.3 |
HSPA13 |
heat shock protein 70kDa family, member 13 |
| chr9_+_71320557 | 0.10 |
ENST00000541509.1 |
PIP5K1B |
phosphatidylinositol-4-phosphate 5-kinase, type I, beta |
| chr16_-_90085824 | 0.10 |
ENST00000002501.6 |
DBNDD1 |
dysbindin (dystrobrevin binding protein 1) domain containing 1 |
| chr6_-_31864977 | 0.10 |
ENST00000395728.3 ENST00000375528.4 |
EHMT2 |
euchromatic histone-lysine N-methyltransferase 2 |
| chr19_-_52097613 | 0.10 |
ENST00000301439.3 |
AC018755.1 |
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr22_+_31742875 | 0.09 |
ENST00000504184.2 |
AC005003.1 |
CDNA FLJ20464 fis, clone KAT06158; HCG1777549; Uncharacterized protein |
| chrX_-_119445263 | 0.09 |
ENST00000309720.5 |
TMEM255A |
transmembrane protein 255A |
| chr5_-_159739532 | 0.09 |
ENST00000520748.1 ENST00000393977.3 ENST00000257536.7 |
CCNJL |
cyclin J-like |
| chr8_+_38644778 | 0.09 |
ENST00000276520.8 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr2_-_32390801 | 0.09 |
ENST00000608489.1 |
RP11-563N4.1 |
RP11-563N4.1 |
| chr1_-_54304212 | 0.09 |
ENST00000540001.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr12_+_54955235 | 0.09 |
ENST00000550620.1 |
PDE1B |
phosphodiesterase 1B, calmodulin-dependent |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.4 | 1.9 | GO:0032449 | CBM complex(GO:0032449) |
| 0.2 | 0.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.2 | 2.7 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.5 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.3 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.6 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.1 | 0.2 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.0 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.2 | GO:0071546 | perinucleolar chromocenter(GO:0010370) pi-body(GO:0071546) |
| 0.0 | 0.5 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 1.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.3 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 2.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 0.7 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.2 | 1.3 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.4 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 1.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 2.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.3 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 3.2 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.5 | GO:0051430 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 3.5 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 4.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 3.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 2.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 1.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.0 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:1904799 | negative regulation of dendrite extension(GO:1903860) regulation of neuron remodeling(GO:1904799) negative regulation of neuron remodeling(GO:1904800) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.6 | 1.9 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.5 | 2.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.4 | 2.6 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 1.0 | GO:0045082 | interleukin-4 biosynthetic process(GO:0042097) positive regulation of interleukin-10 biosynthetic process(GO:0045082) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 0.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.2 | 0.6 | GO:0098904 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) regulation of AV node cell action potential(GO:0098904) regulation of bundle of His cell action potential(GO:0098905) |
| 0.2 | 1.9 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.2 | 0.5 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.4 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.4 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.1 | 0.3 | GO:0021780 | noradrenergic neuron development(GO:0003358) neuroblast differentiation(GO:0014016) spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) stem cell fate determination(GO:0048867) regulation of timing of neuron differentiation(GO:0060164) olfactory pit development(GO:0060166) |
| 0.1 | 1.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.3 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.1 | 0.6 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 1.9 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.5 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.2 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 | 0.3 | GO:0000023 | maltose metabolic process(GO:0000023) diaphragm contraction(GO:0002086) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.3 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.1 | 0.4 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 0.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.1 | 0.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.2 | GO:0042823 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 1.8 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.2 | GO:0070631 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.2 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.1 | 1.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.5 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.3 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.3 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 1.0 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.7 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.8 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 1.6 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.7 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.6 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.4 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.0 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.4 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.6 | GO:0061098 | positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 1.2 | GO:0030516 | regulation of axon extension(GO:0030516) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.2 | GO:0090051 | negative regulation of cell migration involved in sprouting angiogenesis(GO:0090051) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.9 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.6 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |