Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
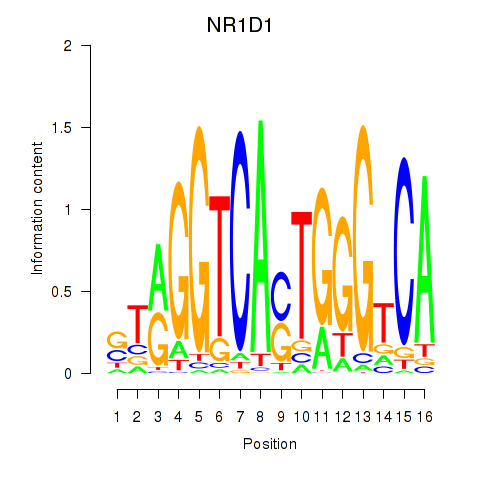
Results for NR1D1
Z-value: 0.59
Transcription factors associated with NR1D1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1D1
|
ENSG00000126368.5 | NR1D1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1D1 | hg19_v2_chr17_-_38256973_38256990 | 0.44 | 8.6e-02 | Click! |
Activity profile of NR1D1 motif
Sorted Z-values of NR1D1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1D1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_46740730 | 0.95 |
ENST00000311907.5 ENST00000530231.1 ENST00000442468.1 |
F2 |
coagulation factor II (thrombin) |
| chr9_+_103790991 | 0.89 |
ENST00000374874.3 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr17_+_79968655 | 0.78 |
ENST00000583744.1 |
ASPSCR1 |
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr11_+_7534999 | 0.74 |
ENST00000528947.1 ENST00000299492.4 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr8_+_11561660 | 0.72 |
ENST00000526716.1 ENST00000335135.4 ENST00000528027.1 |
GATA4 |
GATA binding protein 4 |
| chr2_-_64371546 | 0.64 |
ENST00000358912.4 |
PELI1 |
pellino E3 ubiquitin protein ligase 1 |
| chr18_-_19284724 | 0.57 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr6_+_31543334 | 0.54 |
ENST00000449264.2 |
TNF |
tumor necrosis factor |
| chr1_+_20915409 | 0.49 |
ENST00000375071.3 |
CDA |
cytidine deaminase |
| chr17_-_76128488 | 0.47 |
ENST00000322914.3 |
TMC6 |
transmembrane channel-like 6 |
| chr19_-_23578220 | 0.44 |
ENST00000595533.1 ENST00000397082.2 ENST00000599743.1 ENST00000300619.7 |
ZNF91 |
zinc finger protein 91 |
| chr17_-_3599327 | 0.43 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chrX_+_68725084 | 0.39 |
ENST00000252338.4 |
FAM155B |
family with sequence similarity 155, member B |
| chr13_+_113622757 | 0.38 |
ENST00000375604.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr1_+_44398943 | 0.38 |
ENST00000372359.5 ENST00000414809.3 |
ARTN |
artemin |
| chr17_-_3599492 | 0.38 |
ENST00000435558.1 ENST00000345901.3 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr19_+_21324827 | 0.38 |
ENST00000600692.1 ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431 |
zinc finger protein 431 |
| chr11_+_125365110 | 0.37 |
ENST00000527818.1 |
AP000708.1 |
AP000708.1 |
| chr1_-_147245484 | 0.32 |
ENST00000271348.2 |
GJA5 |
gap junction protein, alpha 5, 40kDa |
| chr13_+_113622810 | 0.31 |
ENST00000397030.1 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr3_+_58223228 | 0.29 |
ENST00000478253.1 ENST00000295962.4 |
ABHD6 |
abhydrolase domain containing 6 |
| chr16_+_67233007 | 0.29 |
ENST00000360833.1 ENST00000393997.2 |
ELMO3 |
engulfment and cell motility 3 |
| chr17_-_38074842 | 0.29 |
ENST00000309481.7 |
GSDMB |
gasdermin B |
| chr17_-_38074859 | 0.28 |
ENST00000520542.1 ENST00000418519.1 ENST00000394179.1 |
GSDMB |
gasdermin B |
| chr16_+_67233412 | 0.27 |
ENST00000477898.1 |
ELMO3 |
engulfment and cell motility 3 |
| chr7_+_64126535 | 0.27 |
ENST00000344930.3 |
ZNF107 |
zinc finger protein 107 |
| chr14_-_23770683 | 0.27 |
ENST00000561437.1 ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E |
protein phosphatase 1, regulatory subunit 3E |
| chr7_+_94139105 | 0.26 |
ENST00000297273.4 |
CASD1 |
CAS1 domain containing 1 |
| chr6_+_167525277 | 0.26 |
ENST00000400926.2 |
CCR6 |
chemokine (C-C motif) receptor 6 |
| chr9_+_126773880 | 0.25 |
ENST00000373615.4 |
LHX2 |
LIM homeobox 2 |
| chr17_-_79139817 | 0.25 |
ENST00000326724.4 |
AATK |
apoptosis-associated tyrosine kinase |
| chr19_-_14629224 | 0.23 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr22_+_37415700 | 0.21 |
ENST00000397129.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr22_+_37415728 | 0.20 |
ENST00000404802.3 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr2_+_63816295 | 0.20 |
ENST00000539945.1 ENST00000544381.1 |
MDH1 |
malate dehydrogenase 1, NAD (soluble) |
| chr19_+_21264980 | 0.19 |
ENST00000596053.1 ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714 |
zinc finger protein 714 |
| chr5_-_43313574 | 0.19 |
ENST00000325110.6 ENST00000433297.2 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr7_-_128694927 | 0.19 |
ENST00000471166.1 ENST00000265388.5 |
TNPO3 |
transportin 3 |
| chrX_+_24167746 | 0.18 |
ENST00000428571.1 ENST00000539115.1 |
ZFX |
zinc finger protein, X-linked |
| chr19_-_23433144 | 0.18 |
ENST00000418100.1 ENST00000597537.1 ENST00000597037.1 |
ZNF724P |
zinc finger protein 724, pseudogene |
| chr22_+_45898712 | 0.17 |
ENST00000455233.1 ENST00000348697.2 ENST00000402984.3 ENST00000262722.7 ENST00000327858.6 ENST00000442170.2 ENST00000340923.5 ENST00000439835.1 |
FBLN1 |
fibulin 1 |
| chr22_+_37415776 | 0.17 |
ENST00000341116.3 ENST00000429360.2 ENST00000404393.1 |
MPST |
mercaptopyruvate sulfurtransferase |
| chr3_+_171758344 | 0.16 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chr1_+_168545711 | 0.16 |
ENST00000367818.3 |
XCL1 |
chemokine (C motif) ligand 1 |
| chr19_+_22235310 | 0.16 |
ENST00000600162.1 |
ZNF257 |
zinc finger protein 257 |
| chr7_-_100493744 | 0.16 |
ENST00000428317.1 ENST00000441605.1 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr1_-_168513229 | 0.15 |
ENST00000367819.2 |
XCL2 |
chemokine (C motif) ligand 2 |
| chr16_-_18468926 | 0.15 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chrX_+_24167828 | 0.14 |
ENST00000379188.3 ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX |
zinc finger protein, X-linked |
| chr19_-_38806560 | 0.13 |
ENST00000591755.1 ENST00000337679.8 ENST00000339413.6 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
| chr3_-_48723268 | 0.13 |
ENST00000439518.1 ENST00000416649.2 ENST00000341520.4 ENST00000294129.2 |
NCKIPSD |
NCK interacting protein with SH3 domain |
| chr8_+_105235572 | 0.13 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_-_38256973 | 0.13 |
ENST00000246672.3 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
| chr1_-_11865982 | 0.12 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr2_+_63816087 | 0.12 |
ENST00000409908.1 ENST00000442225.1 ENST00000409476.1 ENST00000436321.1 |
MDH1 |
malate dehydrogenase 1, NAD (soluble) |
| chr2_-_89513402 | 0.12 |
ENST00000498435.1 |
IGKV1-27 |
immunoglobulin kappa variable 1-27 |
| chr1_+_2398876 | 0.12 |
ENST00000449969.1 |
PLCH2 |
phospholipase C, eta 2 |
| chr2_-_103353277 | 0.11 |
ENST00000258436.5 |
MFSD9 |
major facilitator superfamily domain containing 9 |
| chr17_+_6899366 | 0.11 |
ENST00000251535.6 |
ALOX12 |
arachidonate 12-lipoxygenase |
| chr1_-_111991850 | 0.11 |
ENST00000411751.2 |
WDR77 |
WD repeat domain 77 |
| chr15_-_81282133 | 0.11 |
ENST00000261758.4 |
MESDC2 |
mesoderm development candidate 2 |
| chr7_-_142630820 | 0.11 |
ENST00000442623.1 ENST00000265310.1 |
TRPV5 |
transient receptor potential cation channel, subfamily V, member 5 |
| chrY_+_2803322 | 0.10 |
ENST00000383052.1 ENST00000155093.3 ENST00000449237.1 ENST00000443793.1 |
ZFY |
zinc finger protein, Y-linked |
| chr7_-_100493482 | 0.10 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr1_-_11866034 | 0.10 |
ENST00000376590.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr2_+_113342163 | 0.10 |
ENST00000409719.1 |
CHCHD5 |
coiled-coil-helix-coiled-coil-helix domain containing 5 |
| chr4_+_1873100 | 0.09 |
ENST00000508803.1 |
WHSC1 |
Wolf-Hirschhorn syndrome candidate 1 |
| chr7_-_128695147 | 0.09 |
ENST00000482320.1 ENST00000393245.1 ENST00000471234.1 |
TNPO3 |
transportin 3 |
| chr2_-_89442621 | 0.09 |
ENST00000492167.1 |
IGKV3-20 |
immunoglobulin kappa variable 3-20 |
| chr1_-_11865351 | 0.09 |
ENST00000413656.1 ENST00000376585.1 ENST00000423400.1 ENST00000431243.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr1_+_168250194 | 0.09 |
ENST00000367821.3 |
TBX19 |
T-box 19 |
| chr19_-_12833164 | 0.08 |
ENST00000356861.5 |
TNPO2 |
transportin 2 |
| chr14_+_74815116 | 0.08 |
ENST00000256362.4 |
VRTN |
vertebrae development associated |
| chr7_+_97736197 | 0.07 |
ENST00000297293.5 |
LMTK2 |
lemur tyrosine kinase 2 |
| chr13_+_100741269 | 0.06 |
ENST00000376286.4 ENST00000376279.3 ENST00000376285.1 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
| chr2_-_73053126 | 0.06 |
ENST00000272427.6 ENST00000410104.1 |
EXOC6B |
exocyst complex component 6B |
| chr7_+_139025875 | 0.06 |
ENST00000297534.6 |
C7orf55 |
chromosome 7 open reading frame 55 |
| chr1_+_203764742 | 0.06 |
ENST00000432282.1 ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A |
zinc finger CCCH-type containing 11A |
| chr1_+_218519577 | 0.06 |
ENST00000366930.4 ENST00000366929.4 |
TGFB2 |
transforming growth factor, beta 2 |
| chr17_-_36347030 | 0.05 |
ENST00000518551.1 |
TBC1D3 |
TBC1 domain family, member 3 |
| chr19_-_22605136 | 0.05 |
ENST00000357774.5 ENST00000601553.1 ENST00000593657.1 |
ZNF98 |
zinc finger protein 98 |
| chr16_+_1832902 | 0.05 |
ENST00000262302.9 ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2 |
nucleotide binding protein 2 |
| chr2_+_90153696 | 0.03 |
ENST00000417279.2 |
IGKV3D-15 |
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr1_+_197382957 | 0.03 |
ENST00000367397.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr19_-_12833361 | 0.03 |
ENST00000592287.1 |
TNPO2 |
transportin 2 |
| chr19_+_21265028 | 0.03 |
ENST00000291770.7 |
ZNF714 |
zinc finger protein 714 |
| chr1_-_156918806 | 0.03 |
ENST00000315174.8 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr1_+_110210644 | 0.03 |
ENST00000369831.2 ENST00000442650.1 ENST00000369827.3 ENST00000460717.3 ENST00000241337.4 ENST00000467579.3 ENST00000414179.2 ENST00000369829.2 |
GSTM2 |
glutathione S-transferase mu 2 (muscle) |
| chr19_-_52035044 | 0.02 |
ENST00000359982.4 ENST00000436458.1 ENST00000425629.3 ENST00000391797.3 ENST00000343300.4 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr16_-_70712229 | 0.02 |
ENST00000562883.2 |
MTSS1L |
metastasis suppressor 1-like |
| chr2_+_203776937 | 0.01 |
ENST00000402905.3 ENST00000414490.1 ENST00000431787.1 ENST00000444724.1 ENST00000414857.1 ENST00000430899.1 ENST00000445120.1 ENST00000441569.1 ENST00000432024.1 ENST00000443740.1 ENST00000414439.1 ENST00000428585.1 ENST00000545253.1 ENST00000545262.1 ENST00000447539.1 ENST00000456821.2 ENST00000434998.1 ENST00000320443.8 |
CARF |
calcium responsive transcription factor |
| chr16_+_71392616 | 0.01 |
ENST00000349553.5 ENST00000302628.4 ENST00000562305.1 |
CALB2 |
calbindin 2 |
| chr19_-_19932501 | 0.01 |
ENST00000540806.2 ENST00000590766.1 ENST00000587452.1 ENST00000545006.1 ENST00000590319.1 ENST00000587461.1 ENST00000450683.2 ENST00000443905.2 ENST00000590274.1 |
ZNF506 CTC-559E9.4 |
zinc finger protein 506 CTC-559E9.4 |
| chr6_-_31938700 | 0.01 |
ENST00000495340.1 |
DXO |
decapping exoribonuclease |
| chr1_-_41131326 | 0.01 |
ENST00000372684.3 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
| chr19_+_21203426 | 0.00 |
ENST00000261560.5 ENST00000599548.1 ENST00000594110.1 |
ZNF430 |
zinc finger protein 430 |
| chr8_+_101162812 | 0.00 |
ENST00000353107.3 ENST00000522439.1 |
POLR2K |
polymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0071751 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.2 | 0.6 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.2 | 0.9 | GO:0051801 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.2 | 0.5 | GO:0019858 | cytosine metabolic process(GO:0019858) |
| 0.2 | 0.6 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.6 | GO:0009439 | cyanate metabolic process(GO:0009439) cyanate catabolic process(GO:0009440) |
| 0.1 | 0.3 | GO:0098904 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) regulation of AV node cell action potential(GO:0098904) regulation of bundle of His cell action potential(GO:0098905) |
| 0.1 | 0.3 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.3 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.4 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.3 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:2000412 | positive regulation of thymocyte migration(GO:2000412) positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.3 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:2001303 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.8 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.3 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.6 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.1 | 0.4 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.1 | 0.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.8 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.5 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |