Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
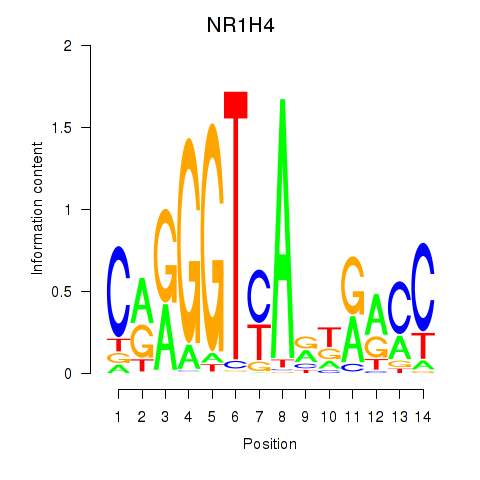
Results for NR1H4
Z-value: 0.53
Transcription factors associated with NR1H4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H4
|
ENSG00000012504.9 | NR1H4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1H4 | hg19_v2_chr12_+_100867694_100867706 | -0.83 | 8.4e-05 | Click! |
Activity profile of NR1H4 motif
Sorted Z-values of NR1H4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1H4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_33172221 | 1.24 |
ENST00000354476.3 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33172012 | 1.20 |
ENST00000404816.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr13_-_38172863 | 1.01 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr9_-_117853297 | 0.93 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr17_+_45286387 | 0.75 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_-_6341724 | 0.68 |
ENST00000530979.1 |
PRKCDBP |
protein kinase C, delta binding protein |
| chr5_+_131593364 | 0.66 |
ENST00000253754.3 ENST00000379018.3 |
PDLIM4 |
PDZ and LIM domain 4 |
| chr8_+_9046503 | 0.61 |
ENST00000512942.2 |
RP11-10A14.5 |
RP11-10A14.5 |
| chr8_-_23261589 | 0.60 |
ENST00000524168.1 ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2 |
lysyl oxidase-like 2 |
| chr9_-_35685452 | 0.60 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr3_-_38071122 | 0.51 |
ENST00000334661.4 |
PLCD1 |
phospholipase C, delta 1 |
| chr10_-_49701686 | 0.49 |
ENST00000417247.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr2_-_106015527 | 0.48 |
ENST00000344213.4 ENST00000358129.4 |
FHL2 |
four and a half LIM domains 2 |
| chr10_+_105726862 | 0.46 |
ENST00000335753.4 ENST00000369755.3 |
SLK |
STE20-like kinase |
| chr1_-_36789755 | 0.45 |
ENST00000270824.1 |
EVA1B |
eva-1 homolog B (C. elegans) |
| chrX_-_106449656 | 0.44 |
ENST00000372466.4 ENST00000421752.1 ENST00000372461.3 |
NUP62CL |
nucleoporin 62kDa C-terminal like |
| chr1_-_25747283 | 0.43 |
ENST00000346452.4 ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE |
Rh blood group, CcEe antigens |
| chr5_+_32710736 | 0.43 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr14_-_21492251 | 0.41 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr11_-_64511575 | 0.40 |
ENST00000431822.1 ENST00000377486.3 ENST00000394432.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr22_+_20105012 | 0.39 |
ENST00000331821.3 ENST00000411892.1 |
RANBP1 |
RAN binding protein 1 |
| chr1_-_109584608 | 0.38 |
ENST00000400794.3 ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47 |
WD repeat domain 47 |
| chr1_-_109584768 | 0.38 |
ENST00000357672.3 |
WDR47 |
WD repeat domain 47 |
| chr11_-_10879572 | 0.38 |
ENST00000413761.2 |
ZBED5 |
zinc finger, BED-type containing 5 |
| chr14_-_21492113 | 0.37 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr1_-_109584716 | 0.37 |
ENST00000531337.1 ENST00000529074.1 ENST00000369965.4 |
WDR47 |
WD repeat domain 47 |
| chr2_-_85637459 | 0.36 |
ENST00000409921.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr6_-_106773610 | 0.36 |
ENST00000369076.3 ENST00000369070.1 |
ATG5 |
autophagy related 5 |
| chr11_-_10879593 | 0.35 |
ENST00000528289.1 ENST00000432999.2 |
ZBED5 |
zinc finger, BED-type containing 5 |
| chr22_+_20105259 | 0.35 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr15_-_75932528 | 0.35 |
ENST00000403490.1 |
IMP3 |
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr1_+_86046433 | 0.34 |
ENST00000451137.2 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
| chr6_-_106773291 | 0.33 |
ENST00000343245.3 |
ATG5 |
autophagy related 5 |
| chr1_+_156024525 | 0.32 |
ENST00000368305.4 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr1_+_156024552 | 0.32 |
ENST00000368304.5 ENST00000368302.3 |
LAMTOR2 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr3_+_54157480 | 0.31 |
ENST00000490478.1 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr11_-_33795893 | 0.31 |
ENST00000526785.1 ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3 |
F-box protein 3 |
| chr16_+_23847339 | 0.30 |
ENST00000303531.7 |
PRKCB |
protein kinase C, beta |
| chr12_+_58013693 | 0.30 |
ENST00000320442.4 ENST00000379218.2 |
SLC26A10 |
solute carrier family 26, member 10 |
| chr15_+_83776137 | 0.29 |
ENST00000322019.9 |
TM6SF1 |
transmembrane 6 superfamily member 1 |
| chr7_-_131241361 | 0.28 |
ENST00000378555.3 ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL |
podocalyxin-like |
| chr6_+_127588020 | 0.28 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr4_+_41992489 | 0.27 |
ENST00000264451.7 |
SLC30A9 |
solute carrier family 30 (zinc transporter), member 9 |
| chr1_+_25599018 | 0.27 |
ENST00000417538.2 ENST00000357542.4 ENST00000568195.1 ENST00000342055.5 ENST00000423810.2 |
RHD |
Rh blood group, D antigen |
| chr12_+_100661156 | 0.27 |
ENST00000360820.2 |
SCYL2 |
SCY1-like 2 (S. cerevisiae) |
| chr8_-_131028869 | 0.27 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr1_+_25598872 | 0.27 |
ENST00000328664.4 |
RHD |
Rh blood group, D antigen |
| chr8_-_131028660 | 0.26 |
ENST00000401979.2 ENST00000517654.1 ENST00000522361.1 ENST00000518167.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr1_+_25598989 | 0.26 |
ENST00000454452.2 |
RHD |
Rh blood group, D antigen |
| chrX_+_66764375 | 0.25 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr6_-_106773491 | 0.24 |
ENST00000360666.4 |
ATG5 |
autophagy related 5 |
| chr5_-_139944196 | 0.23 |
ENST00000357560.4 |
APBB3 |
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr3_+_50606901 | 0.22 |
ENST00000455834.1 |
HEMK1 |
HemK methyltransferase family member 1 |
| chr11_-_58345569 | 0.22 |
ENST00000528954.1 ENST00000528489.1 |
LPXN |
leupaxin |
| chr22_-_20104700 | 0.22 |
ENST00000439169.2 ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A |
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr1_+_26560676 | 0.22 |
ENST00000451429.2 ENST00000252992.4 |
CEP85 |
centrosomal protein 85kDa |
| chr17_-_53809473 | 0.21 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr6_+_127587755 | 0.21 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr17_+_59529743 | 0.21 |
ENST00000589003.1 ENST00000393853.4 |
TBX4 |
T-box 4 |
| chr12_+_6898638 | 0.21 |
ENST00000011653.4 |
CD4 |
CD4 molecule |
| chr16_+_23847267 | 0.21 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr5_+_162930114 | 0.21 |
ENST00000280969.5 |
MAT2B |
methionine adenosyltransferase II, beta |
| chr3_-_123512688 | 0.21 |
ENST00000475616.1 |
MYLK |
myosin light chain kinase |
| chr3_+_49058444 | 0.20 |
ENST00000326925.6 ENST00000395458.2 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr10_-_75634326 | 0.20 |
ENST00000322635.3 ENST00000444854.2 ENST00000423381.1 ENST00000322680.3 ENST00000394762.2 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr5_+_149569520 | 0.20 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr19_+_10196981 | 0.20 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr17_+_73257742 | 0.20 |
ENST00000579761.1 ENST00000245539.6 |
MRPS7 |
mitochondrial ribosomal protein S7 |
| chr10_-_75634260 | 0.20 |
ENST00000372765.1 ENST00000351293.3 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_-_149908217 | 0.20 |
ENST00000369140.3 |
MTMR11 |
myotubularin related protein 11 |
| chr19_+_1026298 | 0.20 |
ENST00000263097.4 |
CNN2 |
calponin 2 |
| chrX_+_47420516 | 0.19 |
ENST00000377045.4 ENST00000290277.6 ENST00000377039.2 |
ARAF |
v-raf murine sarcoma 3611 viral oncogene homolog |
| chr17_-_4852332 | 0.19 |
ENST00000572383.1 |
PFN1 |
profilin 1 |
| chr19_+_10197463 | 0.19 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr5_-_139943830 | 0.19 |
ENST00000412920.3 ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3 |
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr19_+_1026566 | 0.19 |
ENST00000348419.3 ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2 |
calponin 2 |
| chr9_+_74526384 | 0.19 |
ENST00000334731.2 ENST00000377031.3 |
C9orf85 |
chromosome 9 open reading frame 85 |
| chr19_-_51220176 | 0.18 |
ENST00000359082.3 ENST00000293441.1 |
SHANK1 |
SH3 and multiple ankyrin repeat domains 1 |
| chr5_+_140579162 | 0.18 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr19_-_50311896 | 0.18 |
ENST00000529634.2 |
FUZ |
fuzzy planar cell polarity protein |
| chr12_+_123874589 | 0.17 |
ENST00000437502.1 |
SETD8 |
SET domain containing (lysine methyltransferase) 8 |
| chr11_-_57298187 | 0.17 |
ENST00000525158.1 ENST00000257245.4 ENST00000525587.1 |
TIMM10 |
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr17_+_73257945 | 0.17 |
ENST00000579002.1 |
MRPS7 |
mitochondrial ribosomal protein S7 |
| chr5_-_137878887 | 0.17 |
ENST00000507939.1 ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1 |
eukaryotic translation termination factor 1 |
| chr3_-_120461378 | 0.17 |
ENST00000273375.3 |
RABL3 |
RAB, member of RAS oncogene family-like 3 |
| chr14_-_21945057 | 0.16 |
ENST00000397762.1 |
RAB2B |
RAB2B, member RAS oncogene family |
| chr9_-_138391692 | 0.15 |
ENST00000429260.2 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr7_+_99102573 | 0.15 |
ENST00000394170.2 |
ZKSCAN5 |
zinc finger with KRAB and SCAN domains 5 |
| chr14_+_101297740 | 0.15 |
ENST00000555928.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr12_+_67663056 | 0.15 |
ENST00000545606.1 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
| chr11_+_64073699 | 0.14 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr11_-_4414880 | 0.14 |
ENST00000254436.7 ENST00000543625.1 |
TRIM21 |
tripartite motif containing 21 |
| chr2_-_182545603 | 0.14 |
ENST00000295108.3 |
NEUROD1 |
neuronal differentiation 1 |
| chr3_+_52489503 | 0.14 |
ENST00000345716.4 |
NISCH |
nischarin |
| chr3_-_28390581 | 0.13 |
ENST00000479665.1 |
AZI2 |
5-azacytidine induced 2 |
| chr9_-_35958151 | 0.13 |
ENST00000341959.2 |
OR2S2 |
olfactory receptor, family 2, subfamily S, member 2 |
| chr12_+_16035307 | 0.12 |
ENST00000538352.1 ENST00000025399.6 ENST00000419869.2 |
STRAP |
serine/threonine kinase receptor associated protein |
| chr22_+_20104947 | 0.12 |
ENST00000402752.1 |
RANBP1 |
RAN binding protein 1 |
| chr2_+_192542850 | 0.12 |
ENST00000410026.2 |
NABP1 |
nucleic acid binding protein 1 |
| chr6_+_30594619 | 0.12 |
ENST00000318999.7 ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
| chr3_-_126327398 | 0.12 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr5_-_16509101 | 0.11 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr22_+_31090793 | 0.11 |
ENST00000332585.6 ENST00000382310.3 ENST00000446658.2 |
OSBP2 |
oxysterol binding protein 2 |
| chr1_-_16939976 | 0.11 |
ENST00000430580.2 |
NBPF1 |
neuroblastoma breakpoint family, member 1 |
| chr9_+_138391805 | 0.11 |
ENST00000371785.1 |
MRPS2 |
mitochondrial ribosomal protein S2 |
| chr1_-_37499726 | 0.11 |
ENST00000373091.3 ENST00000373093.4 |
GRIK3 |
glutamate receptor, ionotropic, kainate 3 |
| chr11_-_72070206 | 0.11 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr11_+_67798114 | 0.10 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr1_+_21766641 | 0.10 |
ENST00000342104.5 |
NBPF3 |
neuroblastoma breakpoint family, member 3 |
| chr19_+_41117770 | 0.10 |
ENST00000601032.1 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr11_-_47664072 | 0.09 |
ENST00000542981.1 ENST00000530428.1 ENST00000302503.3 |
MTCH2 |
mitochondrial carrier 2 |
| chr17_-_43568062 | 0.09 |
ENST00000421073.2 ENST00000584420.1 ENST00000589780.1 ENST00000430334.3 |
PLEKHM1 |
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr11_+_45868957 | 0.09 |
ENST00000443527.2 |
CRY2 |
cryptochrome 2 (photolyase-like) |
| chr17_-_59940830 | 0.09 |
ENST00000259008.2 |
BRIP1 |
BRCA1 interacting protein C-terminal helicase 1 |
| chr6_-_33771757 | 0.09 |
ENST00000507738.1 ENST00000266003.5 ENST00000430124.2 |
MLN |
motilin |
| chr22_-_50700140 | 0.08 |
ENST00000215659.8 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr1_+_152658599 | 0.08 |
ENST00000368780.3 |
LCE2B |
late cornified envelope 2B |
| chr19_-_44860820 | 0.08 |
ENST00000354340.4 ENST00000337401.4 ENST00000587909.1 |
ZNF112 |
zinc finger protein 112 |
| chr14_+_24025194 | 0.08 |
ENST00000404535.3 ENST00000288014.6 |
THTPA |
thiamine triphosphatase |
| chr20_-_3644046 | 0.08 |
ENST00000290417.2 ENST00000319242.3 |
GFRA4 |
GDNF family receptor alpha 4 |
| chr7_-_752074 | 0.08 |
ENST00000360274.4 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
| chr1_+_12123414 | 0.08 |
ENST00000263932.2 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
| chr14_+_24025462 | 0.08 |
ENST00000556015.1 ENST00000554970.1 ENST00000554789.1 |
THTPA |
thiamine triphosphatase |
| chr12_-_6451186 | 0.08 |
ENST00000540022.1 ENST00000536194.1 |
TNFRSF1A |
tumor necrosis factor receptor superfamily, member 1A |
| chr1_+_28206150 | 0.07 |
ENST00000456990.1 |
THEMIS2 |
thymocyte selection associated family member 2 |
| chrX_-_132549506 | 0.07 |
ENST00000370828.3 |
GPC4 |
glypican 4 |
| chr12_-_57914275 | 0.07 |
ENST00000547303.1 ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3 |
DNA-damage-inducible transcript 3 |
| chr12_+_120875910 | 0.07 |
ENST00000551806.1 |
AL021546.6 |
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr3_+_14989186 | 0.07 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr5_-_83680603 | 0.07 |
ENST00000296591.5 |
EDIL3 |
EGF-like repeats and discoidin I-like domains 3 |
| chr6_+_96025341 | 0.07 |
ENST00000369293.1 ENST00000358812.4 |
MANEA |
mannosidase, endo-alpha |
| chr8_+_110346546 | 0.07 |
ENST00000521662.1 ENST00000521688.1 ENST00000520147.1 |
ENY2 |
enhancer of yellow 2 homolog (Drosophila) |
| chr10_-_119806085 | 0.06 |
ENST00000355624.3 |
RAB11FIP2 |
RAB11 family interacting protein 2 (class I) |
| chr17_-_4852243 | 0.06 |
ENST00000225655.5 |
PFN1 |
profilin 1 |
| chrX_+_7137475 | 0.06 |
ENST00000217961.4 |
STS |
steroid sulfatase (microsomal), isozyme S |
| chr19_+_6361841 | 0.06 |
ENST00000596605.1 |
CLPP |
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr14_-_25103388 | 0.06 |
ENST00000526004.1 ENST00000415355.3 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr3_-_15540055 | 0.06 |
ENST00000605797.1 ENST00000435459.2 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chrX_-_49965009 | 0.06 |
ENST00000437370.1 ENST00000376064.3 ENST00000448865.1 |
AKAP4 |
A kinase (PRKA) anchor protein 4 |
| chr17_-_46035187 | 0.05 |
ENST00000300557.2 |
PRR15L |
proline rich 15-like |
| chrX_+_152224766 | 0.05 |
ENST00000370265.4 ENST00000447306.1 |
PNMA3 |
paraneoplastic Ma antigen 3 |
| chr22_-_21387127 | 0.04 |
ENST00000426145.1 |
SLC7A4 |
solute carrier family 7, member 4 |
| chr6_+_152011628 | 0.04 |
ENST00000404742.1 ENST00000440973.1 |
ESR1 |
estrogen receptor 1 |
| chr22_+_30163340 | 0.04 |
ENST00000330029.6 ENST00000401406.3 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr12_+_98987369 | 0.04 |
ENST00000401722.3 ENST00000188376.5 ENST00000228318.3 ENST00000551917.1 ENST00000548046.1 ENST00000552981.1 ENST00000551265.1 ENST00000550695.1 ENST00000547534.1 ENST00000549338.1 ENST00000548847.1 |
SLC25A3 |
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 |
| chr18_-_3880051 | 0.04 |
ENST00000584874.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr19_+_36142147 | 0.04 |
ENST00000590618.1 |
COX6B1 |
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr12_+_52306113 | 0.04 |
ENST00000547400.1 ENST00000550683.1 ENST00000419526.2 |
ACVRL1 |
activin A receptor type II-like 1 |
| chr5_+_138629417 | 0.03 |
ENST00000510056.1 ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3 |
matrin 3 |
| chr1_+_202317815 | 0.03 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_138629337 | 0.03 |
ENST00000394805.3 ENST00000512876.1 ENST00000513678.1 |
MATR3 |
matrin 3 |
| chr12_-_58135903 | 0.03 |
ENST00000257897.3 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr14_-_25103472 | 0.03 |
ENST00000216341.4 ENST00000382542.1 ENST00000382540.1 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr17_-_73257667 | 0.03 |
ENST00000538886.1 ENST00000580799.1 ENST00000351904.7 ENST00000537686.1 |
GGA3 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
| chr3_-_48632593 | 0.03 |
ENST00000454817.1 ENST00000328333.8 |
COL7A1 |
collagen, type VII, alpha 1 |
| chr3_+_111260954 | 0.02 |
ENST00000283285.5 |
CD96 |
CD96 molecule |
| chr12_-_71003568 | 0.02 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr17_-_74023291 | 0.02 |
ENST00000586740.1 |
EVPL |
envoplakin |
| chr16_+_89238149 | 0.02 |
ENST00000289746.2 |
CDH15 |
cadherin 15, type 1, M-cadherin (myotubule) |
| chr10_+_47083454 | 0.02 |
ENST00000374312.1 |
NPY4R |
neuropeptide Y receptor Y4 |
| chr3_+_121903181 | 0.02 |
ENST00000498619.1 |
CASR |
calcium-sensing receptor |
| chr1_-_160040038 | 0.02 |
ENST00000368089.3 |
KCNJ10 |
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr17_+_34136459 | 0.02 |
ENST00000588240.1 ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr19_+_10563567 | 0.02 |
ENST00000344979.3 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr5_-_177423243 | 0.01 |
ENST00000308304.2 |
PROP1 |
PROP paired-like homeobox 1 |
| chr11_-_65624415 | 0.01 |
ENST00000524553.1 ENST00000527344.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr5_+_138629389 | 0.01 |
ENST00000504045.1 ENST00000504311.1 ENST00000502499.1 |
MATR3 |
matrin 3 |
| chr17_+_27181956 | 0.01 |
ENST00000254928.5 ENST00000580917.2 |
ERAL1 |
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr3_-_49158312 | 0.01 |
ENST00000398892.3 ENST00000453664.1 ENST00000398888.2 |
USP19 |
ubiquitin specific peptidase 19 |
| chr2_-_84686552 | 0.01 |
ENST00000393868.2 |
SUCLG1 |
succinate-CoA ligase, alpha subunit |
| chr4_+_26322409 | 0.01 |
ENST00000514807.1 ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_-_30537839 | 0.01 |
ENST00000380412.5 |
ZNF768 |
zinc finger protein 768 |
| chrX_+_148793714 | 0.01 |
ENST00000355220.5 |
MAGEA11 |
melanoma antigen family A, 11 |
| chr22_-_29457832 | 0.01 |
ENST00000216071.4 |
C22orf31 |
chromosome 22 open reading frame 31 |
| chr3_+_111260856 | 0.01 |
ENST00000352690.4 |
CD96 |
CD96 molecule |
| chr17_+_41177220 | 0.01 |
ENST00000587250.2 ENST00000544533.1 |
RND2 |
Rho family GTPase 2 |
| chr6_-_33547975 | 0.01 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr1_+_32042131 | 0.00 |
ENST00000271064.7 ENST00000537531.1 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 0.9 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.3 | 0.9 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 2.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.2 | 0.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 0.9 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.5 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 0.5 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.2 | GO:0060769 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.1 | 0.3 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.2 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.3 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.8 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.2 | GO:2000314 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.3 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.2 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.3 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 1.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.1 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.8 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.4 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.6 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.6 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 3.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 1.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 0.8 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 0.9 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.2 | 0.5 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.9 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.5 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.6 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.2 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 1.2 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0098988 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) kainate selective glutamate receptor activity(GO:0015277) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.1 | 0.6 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 2.4 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.3 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.4 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |