Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
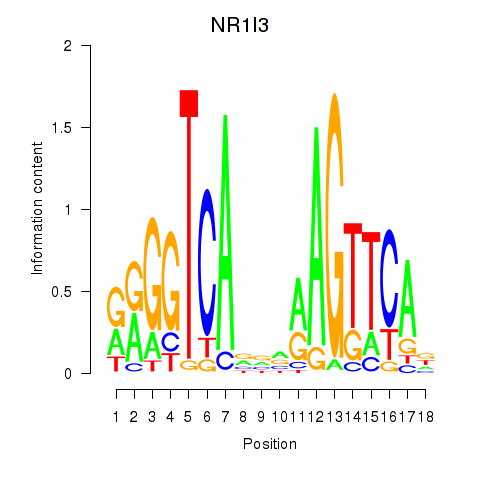
Results for NR1I3
Z-value: 0.45
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | NR1I3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR1I3 | hg19_v2_chr1_-_161207953_161207975 | 0.09 | 7.5e-01 | Click! |
Activity profile of NR1I3 motif
Sorted Z-values of NR1I3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR1I3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_153330322 | 0.68 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr2_-_209118974 | 0.67 |
ENST00000415913.1 ENST00000415282.1 ENST00000446179.1 |
IDH1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr6_+_36097992 | 0.65 |
ENST00000211287.4 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr17_-_26694979 | 0.48 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr17_-_26695013 | 0.47 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr15_+_96876340 | 0.46 |
ENST00000453270.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr10_-_73848764 | 0.38 |
ENST00000317376.4 ENST00000412663.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr1_-_205290865 | 0.37 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr18_-_19283649 | 0.36 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr18_-_19284724 | 0.36 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr16_-_16317321 | 0.34 |
ENST00000205557.7 ENST00000575728.1 |
ABCC6 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 6 |
| chr17_-_53499310 | 0.32 |
ENST00000262065.3 |
MMD |
monocyte to macrophage differentiation-associated |
| chr17_+_32582293 | 0.31 |
ENST00000580907.1 ENST00000225831.4 |
CCL2 |
chemokine (C-C motif) ligand 2 |
| chr4_-_57524061 | 0.30 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr1_+_44401479 | 0.28 |
ENST00000438616.3 |
ARTN |
artemin |
| chr15_+_96875657 | 0.27 |
ENST00000559679.1 ENST00000394171.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr15_+_85523671 | 0.25 |
ENST00000310298.4 ENST00000557957.1 |
PDE8A |
phosphodiesterase 8A |
| chrX_+_128872998 | 0.25 |
ENST00000371106.3 |
XPNPEP2 |
X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound |
| chr11_-_82611448 | 0.24 |
ENST00000393399.2 ENST00000313010.3 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
| chr15_+_43985084 | 0.23 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr15_+_43885252 | 0.21 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr7_+_100770328 | 0.21 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr20_+_62339381 | 0.19 |
ENST00000355969.6 ENST00000357119.4 ENST00000431125.1 ENST00000369967.3 ENST00000328969.5 |
ZGPAT |
zinc finger, CCCH-type with G patch domain |
| chr11_-_104480019 | 0.19 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chr19_-_48894104 | 0.18 |
ENST00000597017.1 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr7_-_31380502 | 0.15 |
ENST00000297142.3 |
NEUROD6 |
neuronal differentiation 6 |
| chr16_-_75018968 | 0.15 |
ENST00000262144.6 |
WDR59 |
WD repeat domain 59 |
| chr2_-_86790593 | 0.14 |
ENST00000263856.4 ENST00000409225.2 |
CHMP3 |
charged multivesicular body protein 3 |
| chr1_+_15250596 | 0.13 |
ENST00000361144.5 |
KAZN |
kazrin, periplakin interacting protein |
| chr13_-_95131923 | 0.13 |
ENST00000377028.5 ENST00000446125.1 |
DCT |
dopachrome tautomerase |
| chr12_+_53693812 | 0.13 |
ENST00000549488.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr1_+_154377669 | 0.12 |
ENST00000368485.3 ENST00000344086.4 |
IL6R |
interleukin 6 receptor |
| chr6_+_108487245 | 0.11 |
ENST00000368986.4 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
| chrX_-_110655391 | 0.11 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chr20_-_44007014 | 0.11 |
ENST00000372726.3 ENST00000537995.1 |
TP53TG5 |
TP53 target 5 |
| chr19_-_46296011 | 0.10 |
ENST00000377735.3 ENST00000270223.6 |
DMWD |
dystrophia myotonica, WD repeat containing |
| chr9_+_34179003 | 0.10 |
ENST00000545103.1 ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1 |
ubiquitin associated protein 1 |
| chr1_+_57320437 | 0.10 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr20_-_45142154 | 0.10 |
ENST00000347606.4 ENST00000457685.2 |
ZNF334 |
zinc finger protein 334 |
| chr19_+_2977444 | 0.09 |
ENST00000246112.4 ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr17_-_3301704 | 0.09 |
ENST00000322608.2 |
OR1E1 |
olfactory receptor, family 1, subfamily E, member 1 |
| chr19_-_49140692 | 0.09 |
ENST00000222122.5 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr17_-_34270668 | 0.08 |
ENST00000293274.4 |
LYZL6 |
lysozyme-like 6 |
| chr3_+_122103014 | 0.07 |
ENST00000232125.5 ENST00000477892.1 ENST00000469967.1 |
FAM162A |
family with sequence similarity 162, member A |
| chr17_-_34270697 | 0.07 |
ENST00000585556.1 |
LYZL6 |
lysozyme-like 6 |
| chrX_+_69674943 | 0.07 |
ENST00000542398.1 |
DLG3 |
discs, large homolog 3 (Drosophila) |
| chr11_-_59950519 | 0.07 |
ENST00000528851.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr16_+_30710462 | 0.07 |
ENST00000262518.4 ENST00000395059.2 ENST00000344771.4 |
SRCAP |
Snf2-related CREBBP activator protein |
| chr1_+_26036093 | 0.06 |
ENST00000374329.1 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr4_-_170897045 | 0.06 |
ENST00000508313.1 |
RP11-205M3.3 |
RP11-205M3.3 |
| chr19_-_51336443 | 0.05 |
ENST00000598673.1 |
KLK15 |
kallikrein-related peptidase 15 |
| chr19_-_49140609 | 0.05 |
ENST00000601104.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr19_-_6604094 | 0.05 |
ENST00000597430.2 |
CD70 |
CD70 molecule |
| chr17_-_62502399 | 0.05 |
ENST00000450599.2 ENST00000585060.1 |
DDX5 |
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_+_145516560 | 0.04 |
ENST00000537888.1 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr2_-_69098566 | 0.04 |
ENST00000295379.1 |
BMP10 |
bone morphogenetic protein 10 |
| chr9_-_80646374 | 0.04 |
ENST00000286548.4 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
| chr17_-_62502639 | 0.04 |
ENST00000225792.5 ENST00000581697.1 ENST00000584279.1 ENST00000577922.1 |
DDX5 |
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr1_+_43803475 | 0.04 |
ENST00000372470.3 ENST00000413998.2 |
MPL |
myeloproliferative leukemia virus oncogene |
| chr14_+_22748980 | 0.03 |
ENST00000390465.2 |
TRAV38-2DV8 |
T cell receptor alpha variable 38-2/delta variable 8 |
| chr1_+_145516252 | 0.03 |
ENST00000369306.3 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr1_-_151119087 | 0.02 |
ENST00000341697.3 ENST00000368914.3 |
SEMA6C |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr6_-_34113856 | 0.02 |
ENST00000538487.2 |
GRM4 |
glutamate receptor, metabotropic 4 |
| chr3_-_50396978 | 0.02 |
ENST00000266025.3 |
TMEM115 |
transmembrane protein 115 |
| chr6_-_31509714 | 0.01 |
ENST00000456662.1 ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr3_-_116164306 | 0.01 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr16_+_577697 | 0.01 |
ENST00000562370.1 ENST00000568988.1 ENST00000219611.2 |
CAPN15 |
calpain 15 |
| chr3_-_58419537 | 0.01 |
ENST00000474765.1 ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB |
pyruvate dehydrogenase (lipoamide) beta |
| chr1_-_32210275 | 0.01 |
ENST00000440175.2 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
| chr1_-_225616515 | 0.01 |
ENST00000338179.2 ENST00000425080.1 |
LBR |
lamin B receptor |
| chr14_-_77787198 | 0.00 |
ENST00000261534.4 |
POMT2 |
protein-O-mannosyltransferase 2 |
| chr11_+_63137251 | 0.00 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr6_+_96969672 | 0.00 |
ENST00000369278.4 |
UFL1 |
UFM1-specific ligase 1 |
| chr9_-_91793675 | 0.00 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.7 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.2 | 0.7 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.7 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.2 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.2 | 0.7 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 0.7 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.3 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.2 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0021847 | ventricular zone neuroblast division(GO:0021847) |
| 0.0 | 0.4 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |