Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
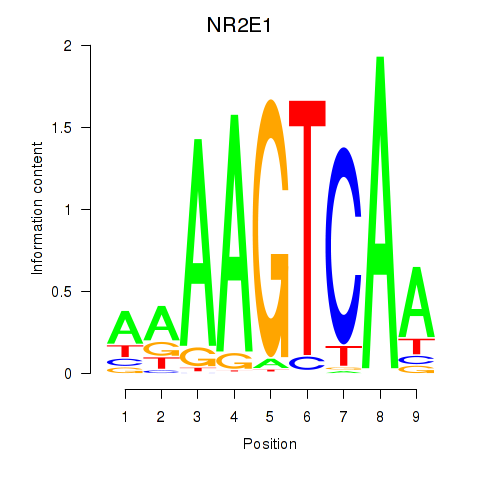
Results for NR2E1
Z-value: 1.05
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | NR2E1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2E1 | hg19_v2_chr6_+_108487245_108487262 | -0.14 | 6.0e-01 | Click! |
Activity profile of NR2E1 motif
Sorted Z-values of NR2E1 motif
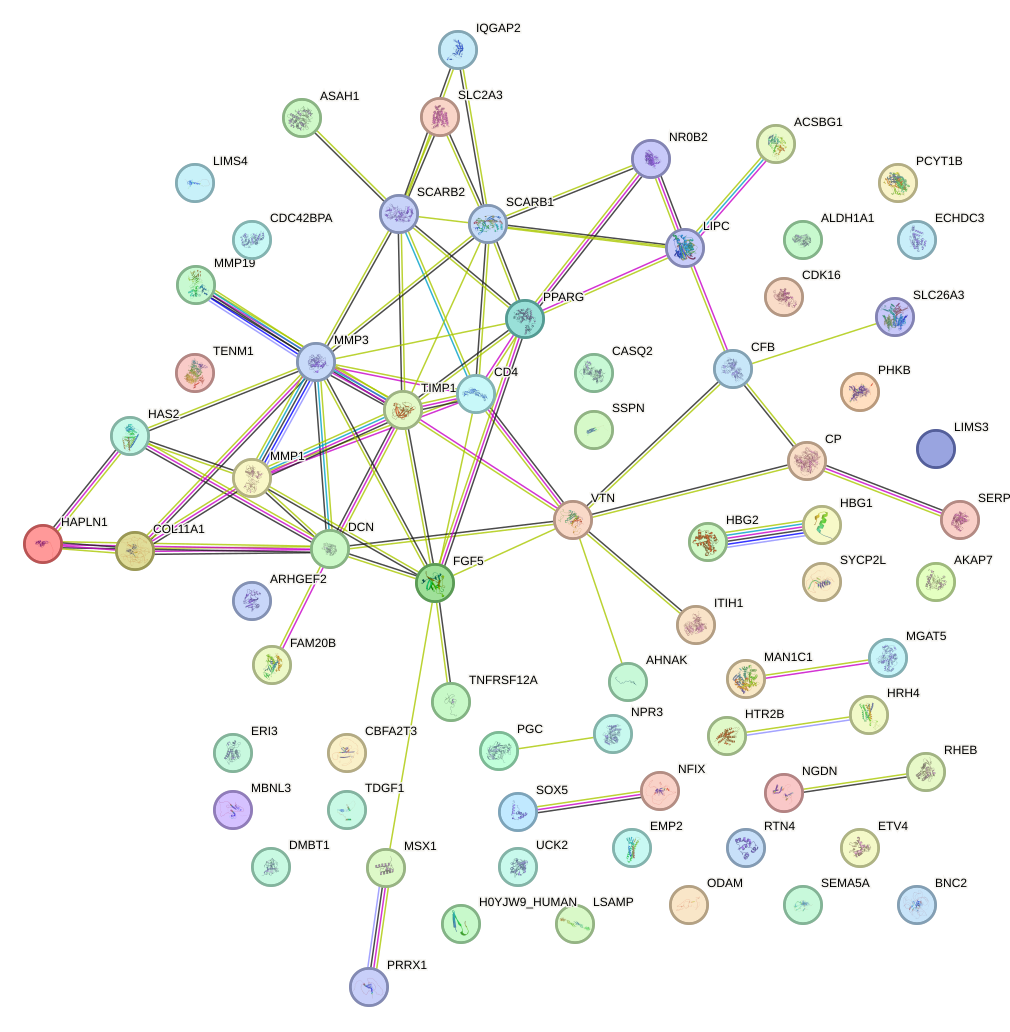
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2E1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91539918 | 3.27 |
ENST00000548218.1 |
DCN |
decorin |
| chr5_-_42811986 | 2.89 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr11_-_5271122 | 2.23 |
ENST00000330597.3 |
HBG1 |
hemoglobin, gamma A |
| chr11_-_5276008 | 2.17 |
ENST00000336906.4 |
HBG2 |
hemoglobin, gamma G |
| chr12_-_91572278 | 1.85 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr11_-_111794446 | 1.67 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr8_-_122653630 | 1.62 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr1_-_103574024 | 1.61 |
ENST00000512756.1 ENST00000370096.3 ENST00000358392.2 ENST00000353414.4 |
COL11A1 |
collagen, type XI, alpha 1 |
| chr12_-_91546926 | 1.37 |
ENST00000550758.1 |
DCN |
decorin |
| chr3_+_69788576 | 1.36 |
ENST00000352241.4 ENST00000448226.2 |
MITF |
microphthalmia-associated transcription factor |
| chr4_+_4861385 | 1.34 |
ENST00000382723.4 |
MSX1 |
msh homeobox 1 |
| chr11_-_111781454 | 1.07 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr3_+_69915385 | 1.05 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr6_-_41715128 | 1.03 |
ENST00000356667.4 ENST00000373025.3 ENST00000425343.2 |
PGC |
progastricsin (pepsinogen C) |
| chr6_+_31916733 | 0.99 |
ENST00000483004.1 |
CFB |
complement factor B |
| chr3_+_69985734 | 0.95 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr3_-_148939835 | 0.94 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr11_-_111781554 | 0.90 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chrX_-_105282712 | 0.90 |
ENST00000372563.1 |
SERPINA7 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr4_-_141348999 | 0.88 |
ENST00000325617.5 |
CLGN |
calmegin |
| chr1_+_25944341 | 0.85 |
ENST00000263979.3 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr12_-_90049878 | 0.77 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr9_-_75567962 | 0.77 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr17_-_66951474 | 0.77 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr3_+_12392971 | 0.75 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr17_-_26695013 | 0.74 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr4_-_111119804 | 0.72 |
ENST00000394607.3 ENST00000302274.3 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
| chr1_-_227505289 | 0.72 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr3_+_69928256 | 0.70 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr10_+_124320156 | 0.70 |
ENST00000338354.3 ENST00000344338.3 ENST00000330163.4 ENST00000368909.3 ENST00000368955.3 ENST00000368956.2 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr12_-_90049828 | 0.69 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr10_+_124320195 | 0.68 |
ENST00000359586.6 |
DMBT1 |
deleted in malignant brain tumors 1 |
| chr20_-_30310693 | 0.67 |
ENST00000307677.4 ENST00000420653.1 |
BCL2L1 |
BCL2-like 1 |
| chr19_+_35940486 | 0.66 |
ENST00000246549.2 |
FFAR2 |
free fatty acid receptor 2 |
| chr16_-_10652993 | 0.66 |
ENST00000536829.1 |
EMP2 |
epithelial membrane protein 2 |
| chr12_+_6898638 | 0.66 |
ENST00000011653.4 |
CD4 |
CD4 molecule |
| chr17_-_26694979 | 0.65 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr1_-_116311402 | 0.64 |
ENST00000261448.5 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
| chr4_+_71063641 | 0.64 |
ENST00000514097.1 |
ODAM |
odontogenic, ameloblast asssociated |
| chr8_-_17942432 | 0.64 |
ENST00000381733.4 ENST00000314146.10 |
ASAH1 |
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr9_-_16727978 | 0.63 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr20_-_30310336 | 0.63 |
ENST00000434194.1 ENST00000376062.2 |
BCL2L1 |
BCL2-like 1 |
| chr20_-_30310656 | 0.63 |
ENST00000376055.4 |
BCL2L1 |
BCL2-like 1 |
| chr3_+_69985792 | 0.58 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chrX_-_128657457 | 0.58 |
ENST00000371121.3 ENST00000371123.1 ENST00000371122.4 |
SMARCA1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr6_+_131521120 | 0.57 |
ENST00000537868.1 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
| chr12_-_8088871 | 0.57 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr15_+_58702742 | 0.57 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chrX_-_114253536 | 0.54 |
ENST00000371936.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chrX_+_47444613 | 0.53 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr5_-_9546180 | 0.53 |
ENST00000382496.5 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr16_+_3068393 | 0.52 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr1_+_178995021 | 0.52 |
ENST00000263733.4 |
FAM20B |
family with sequence similarity 20, member B |
| chr17_-_41623691 | 0.51 |
ENST00000545954.1 |
ETV4 |
ets variant 4 |
| chr11_-_8832182 | 0.50 |
ENST00000527510.1 ENST00000528527.1 ENST00000528523.1 ENST00000313726.6 |
ST5 |
suppression of tumorigenicity 5 |
| chr5_+_32711419 | 0.50 |
ENST00000265074.8 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr11_-_62323702 | 0.49 |
ENST00000530285.1 |
AHNAK |
AHNAK nucleoprotein |
| chr11_-_102714534 | 0.49 |
ENST00000299855.5 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr10_+_11784360 | 0.46 |
ENST00000379215.4 ENST00000420401.1 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
| chr10_+_114709999 | 0.46 |
ENST00000355995.4 ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_26348582 | 0.45 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr1_-_27240455 | 0.45 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr1_+_170633047 | 0.45 |
ENST00000239461.6 ENST00000497230.2 |
PRRX1 |
paired related homeobox 1 |
| chr8_-_86253888 | 0.44 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr2_+_201173667 | 0.43 |
ENST00000409755.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr20_-_22565101 | 0.42 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr4_+_81187753 | 0.42 |
ENST00000312465.7 ENST00000456523.3 |
FGF5 |
fibroblast growth factor 5 |
| chr6_+_125540951 | 0.42 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr3_+_46616017 | 0.41 |
ENST00000542931.1 |
TDGF1 |
teratocarcinoma-derived growth factor 1 |
| chr12_-_56236690 | 0.40 |
ENST00000322569.4 |
MMP19 |
matrix metallopeptidase 19 |
| chr2_-_231989808 | 0.40 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr2_+_192141611 | 0.37 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr2_-_106013400 | 0.36 |
ENST00000409807.1 |
FHL2 |
four and a half LIM domains 2 |
| chr12_-_24102576 | 0.36 |
ENST00000537393.1 ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chrX_-_131547625 | 0.35 |
ENST00000394311.2 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chr8_+_77593474 | 0.35 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr4_-_122085469 | 0.34 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
| chr14_+_22977587 | 0.33 |
ENST00000390504.1 |
TRAJ33 |
T cell receptor alpha joining 33 |
| chr17_-_41623716 | 0.32 |
ENST00000319349.5 |
ETV4 |
ets variant 4 |
| chr5_-_82969405 | 0.31 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chrX_-_131547596 | 0.31 |
ENST00000538204.1 ENST00000370849.3 |
MBNL3 |
muscleblind-like splicing regulator 3 |
| chrX_-_124097620 | 0.31 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr3_-_116164306 | 0.30 |
ENST00000490035.2 |
LSAMP |
limbic system-associated membrane protein |
| chr19_+_13134772 | 0.30 |
ENST00000587760.1 ENST00000585575.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr7_-_151217166 | 0.30 |
ENST00000496004.1 |
RHEB |
Ras homolog enriched in brain |
| chr6_+_10747986 | 0.30 |
ENST00000379542.5 |
TMEM14B |
transmembrane protein 14B |
| chr1_-_44818599 | 0.29 |
ENST00000537474.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr19_-_633576 | 0.29 |
ENST00000588649.2 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
| chr2_+_109237717 | 0.28 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr20_-_30310797 | 0.28 |
ENST00000422920.1 |
BCL2L1 |
BCL2-like 1 |
| chr16_+_47495225 | 0.28 |
ENST00000299167.8 ENST00000323584.5 ENST00000563376.1 |
PHKB |
phosphorylase kinase, beta |
| chrX_-_24690771 | 0.27 |
ENST00000379145.1 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr1_+_165864800 | 0.27 |
ENST00000469256.2 |
UCK2 |
uridine-cytidine kinase 2 |
| chr16_+_47495201 | 0.27 |
ENST00000566044.1 ENST00000455779.1 |
PHKB |
phosphorylase kinase, beta |
| chr7_+_80267973 | 0.26 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr7_-_107443652 | 0.26 |
ENST00000340010.5 ENST00000422236.2 ENST00000453332.1 |
SLC26A3 |
solute carrier family 26 (anion exchanger), member 3 |
| chr10_-_21786179 | 0.26 |
ENST00000377113.5 |
CASC10 |
cancer susceptibility candidate 10 |
| chr2_-_183291741 | 0.25 |
ENST00000351439.5 ENST00000409365.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr1_-_155947951 | 0.25 |
ENST00000313695.7 ENST00000497907.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr13_+_109248500 | 0.25 |
ENST00000356711.2 |
MYO16 |
myosin XVI |
| chr8_+_77593448 | 0.25 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr6_+_10748019 | 0.25 |
ENST00000543878.1 ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L TMEM14B |
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr3_+_52813932 | 0.24 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr2_+_135011731 | 0.24 |
ENST00000281923.2 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr8_+_11561660 | 0.22 |
ENST00000526716.1 ENST00000335135.4 ENST00000528027.1 |
GATA4 |
GATA binding protein 4 |
| chr16_-_73082274 | 0.22 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr3_+_52812523 | 0.22 |
ENST00000540715.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chrX_+_47077632 | 0.21 |
ENST00000457458.2 |
CDK16 |
cyclin-dependent kinase 16 |
| chr14_+_23938891 | 0.21 |
ENST00000408901.3 ENST00000397154.3 ENST00000555128.1 |
NGDN |
neuroguidin, EIF4E binding protein |
| chr16_-_89043377 | 0.21 |
ENST00000436887.2 ENST00000448839.1 ENST00000360302.2 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
| chr19_+_13135386 | 0.21 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr2_-_55237484 | 0.20 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr7_-_151217001 | 0.20 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr5_+_75699040 | 0.20 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr18_+_22040593 | 0.19 |
ENST00000256906.4 |
HRH4 |
histamine receptor H4 |
| chr1_+_161284047 | 0.18 |
ENST00000367975.2 ENST00000342751.4 ENST00000432287.2 ENST00000392169.2 ENST00000513009.1 |
SDHC |
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
| chr9_-_95166841 | 0.18 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr17_-_73775839 | 0.18 |
ENST00000592643.1 ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B |
H3 histone, family 3B (H3.3B) |
| chr5_+_75699149 | 0.18 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr1_-_154155595 | 0.18 |
ENST00000328159.4 ENST00000368531.2 ENST00000323144.7 ENST00000368533.3 ENST00000341372.3 |
TPM3 |
tropomyosin 3 |
| chr1_-_244006528 | 0.17 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr8_+_24241969 | 0.17 |
ENST00000522298.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr12_+_10366016 | 0.17 |
ENST00000546017.1 ENST00000535576.1 ENST00000539170.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr5_-_160973649 | 0.16 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr14_-_71107921 | 0.16 |
ENST00000553982.1 ENST00000500016.1 |
CTD-2540L5.5 CTD-2540L5.6 |
CTD-2540L5.5 CTD-2540L5.6 |
| chr18_+_32290218 | 0.16 |
ENST00000348997.5 ENST00000588949.1 ENST00000597599.1 |
DTNA |
dystrobrevin, alpha |
| chr1_+_212738676 | 0.16 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr8_-_119124045 | 0.16 |
ENST00000378204.2 |
EXT1 |
exostosin glycosyltransferase 1 |
| chr1_-_154155675 | 0.16 |
ENST00000330188.9 ENST00000341485.5 |
TPM3 |
tropomyosin 3 |
| chr17_+_43138664 | 0.16 |
ENST00000258960.2 |
NMT1 |
N-myristoyltransferase 1 |
| chr7_+_39125365 | 0.15 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr5_+_137203465 | 0.14 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr8_+_24241789 | 0.14 |
ENST00000256412.4 ENST00000538205.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr1_+_236558694 | 0.14 |
ENST00000359362.5 |
EDARADD |
EDAR-associated death domain |
| chr5_-_55412774 | 0.14 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr14_+_93651358 | 0.13 |
ENST00000415050.2 |
TMEM251 |
transmembrane protein 251 |
| chr1_-_31661000 | 0.13 |
ENST00000263693.1 ENST00000398657.2 ENST00000526106.1 |
NKAIN1 |
Na+/K+ transporting ATPase interacting 1 |
| chr3_-_116163830 | 0.13 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr9_-_95166884 | 0.12 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr12_-_10324716 | 0.12 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr9_-_95244781 | 0.12 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr1_+_152850787 | 0.12 |
ENST00000368765.3 |
SMCP |
sperm mitochondria-associated cysteine-rich protein |
| chr9_-_21368075 | 0.12 |
ENST00000449498.1 |
IFNA13 |
interferon, alpha 13 |
| chr5_-_142780280 | 0.11 |
ENST00000424646.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr17_-_73975444 | 0.11 |
ENST00000293217.5 ENST00000537812.1 |
ACOX1 |
acyl-CoA oxidase 1, palmitoyl |
| chr1_+_150229554 | 0.11 |
ENST00000369111.4 |
CA14 |
carbonic anhydrase XIV |
| chr18_+_22040620 | 0.11 |
ENST00000426880.2 |
HRH4 |
histamine receptor H4 |
| chr7_-_16505440 | 0.11 |
ENST00000307068.4 |
SOSTDC1 |
sclerostin domain containing 1 |
| chrX_+_47078069 | 0.11 |
ENST00000357227.4 ENST00000519758.1 ENST00000520893.1 ENST00000517426.1 |
CDK16 |
cyclin-dependent kinase 16 |
| chr9_+_21409146 | 0.11 |
ENST00000380205.1 |
IFNA8 |
interferon, alpha 8 |
| chr10_-_73848531 | 0.10 |
ENST00000373109.2 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr9_-_91793675 | 0.10 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr17_-_46667628 | 0.10 |
ENST00000498678.1 |
HOXB3 |
homeobox B3 |
| chr10_-_73848764 | 0.10 |
ENST00000317376.4 ENST00000412663.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr5_+_32711829 | 0.10 |
ENST00000415167.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr14_+_93651296 | 0.10 |
ENST00000283534.4 ENST00000557574.1 |
TMEM251 RP11-371E8.4 |
transmembrane protein 251 Uncharacterized protein |
| chr14_+_39734482 | 0.09 |
ENST00000554392.1 ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5 |
CTAGE family, member 5 |
| chr17_-_56494713 | 0.09 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr17_+_45973516 | 0.09 |
ENST00000376741.4 |
SP2 |
Sp2 transcription factor |
| chr12_-_56236734 | 0.09 |
ENST00000548629.1 |
MMP19 |
matrix metallopeptidase 19 |
| chr1_-_20834586 | 0.09 |
ENST00000264198.3 |
MUL1 |
mitochondrial E3 ubiquitin protein ligase 1 |
| chr3_+_127771212 | 0.09 |
ENST00000243253.3 ENST00000481210.1 |
SEC61A1 |
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr3_+_148508845 | 0.09 |
ENST00000491148.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr17_-_73389854 | 0.09 |
ENST00000578961.1 ENST00000392564.1 ENST00000582582.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr17_-_42327236 | 0.08 |
ENST00000399246.2 |
AC003102.1 |
AC003102.1 |
| chr6_+_26156551 | 0.08 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr6_+_148663729 | 0.08 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr21_-_36260980 | 0.08 |
ENST00000344691.4 ENST00000358356.5 |
RUNX1 |
runt-related transcription factor 1 |
| chr17_-_46667594 | 0.08 |
ENST00000476342.1 ENST00000460160.1 ENST00000472863.1 |
HOXB3 |
homeobox B3 |
| chr6_+_42531798 | 0.08 |
ENST00000372903.2 ENST00000372899.1 ENST00000372901.1 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr2_+_113885138 | 0.08 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr7_-_14026123 | 0.08 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr16_-_18911366 | 0.07 |
ENST00000565224.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr7_+_148936732 | 0.07 |
ENST00000335870.2 |
ZNF212 |
zinc finger protein 212 |
| chr14_+_31494672 | 0.07 |
ENST00000542754.2 ENST00000313566.6 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr9_+_5890802 | 0.07 |
ENST00000381477.3 ENST00000381476.1 ENST00000381471.1 |
MLANA |
melan-A |
| chr8_+_119294456 | 0.06 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chr2_+_198365095 | 0.06 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr14_+_31494841 | 0.06 |
ENST00000556232.1 ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr17_-_73389737 | 0.06 |
ENST00000392563.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr5_-_179047881 | 0.06 |
ENST00000521173.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr9_-_110251836 | 0.06 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr11_-_128457446 | 0.06 |
ENST00000392668.4 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr12_-_58220078 | 0.06 |
ENST00000549039.1 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr1_-_154150651 | 0.06 |
ENST00000302206.5 |
TPM3 |
tropomyosin 3 |
| chr17_+_1959369 | 0.05 |
ENST00000576444.1 ENST00000322941.3 |
HIC1 |
hypermethylated in cancer 1 |
| chr11_-_66104237 | 0.05 |
ENST00000530056.1 |
RIN1 |
Ras and Rab interactor 1 |
| chr17_-_43138463 | 0.05 |
ENST00000310604.4 |
DCAKD |
dephospho-CoA kinase domain containing |
| chr5_-_158757895 | 0.05 |
ENST00000231228.2 |
IL12B |
interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) |
| chr20_+_62369623 | 0.05 |
ENST00000467211.1 |
RP4-583P15.14 |
RP4-583P15.14 |
| chr2_-_176046391 | 0.05 |
ENST00000392541.3 ENST00000409194.1 |
ATP5G3 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr17_+_3379284 | 0.05 |
ENST00000263080.2 |
ASPA |
aspartoacylase |
| chr7_+_100026406 | 0.05 |
ENST00000414441.1 |
MEPCE |
methylphosphate capping enzyme |
| chr4_+_88571429 | 0.05 |
ENST00000339673.6 ENST00000282479.7 |
DMP1 |
dentin matrix acidic phosphoprotein 1 |
| chr16_+_3451184 | 0.05 |
ENST00000575752.1 ENST00000571936.1 ENST00000344823.5 |
ZNF174 |
zinc finger protein 174 |
| chr4_-_108641608 | 0.05 |
ENST00000265174.4 |
PAPSS1 |
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr6_-_108582168 | 0.05 |
ENST00000426155.2 |
SNX3 |
sorting nexin 3 |
| chr4_-_123542224 | 0.05 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr8_+_24298597 | 0.04 |
ENST00000380789.1 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr6_+_107077435 | 0.04 |
ENST00000369046.4 |
QRSL1 |
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 5.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 1.6 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 1.3 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.5 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 4.4 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.2 | 3.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.2 | 2.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 0.7 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.2 | 0.6 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 1.6 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0045281 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.7 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 4.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.2 | 2.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.6 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 0.6 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.7 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 3.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 8.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.8 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.1 | 0.7 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 4.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.7 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.6 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.5 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.2 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.9 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.2 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.6 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.4 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 1.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.7 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 2.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 2.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 1.3 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.3 | 1.6 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.3 | 2.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.3 | 4.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.3 | 1.6 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 0.8 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.7 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.2 | 3.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.6 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.7 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 1.0 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 0.7 | GO:0071350 | interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-15(GO:0071350) |
| 0.2 | 0.6 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.8 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.1 | 0.4 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 1.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.7 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.2 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.1 | 0.4 | GO:0055014 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.1 | 4.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.1 | 0.8 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.1 | 0.9 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.3 | GO:0046035 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.5 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.1 | 0.6 | GO:0086029 | Purkinje myocyte to ventricular cardiac muscle cell signaling(GO:0086029) Purkinje myocyte to ventricular cardiac muscle cell communication(GO:0086068) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.6 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.2 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.5 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.7 | GO:0003093 | regulation of glomerular filtration(GO:0003093) |
| 0.0 | 0.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) positive regulation of axon extension involved in axon guidance(GO:0048842) negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.6 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.3 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.5 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.0 | 0.4 | GO:0007350 | blastoderm segmentation(GO:0007350) tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.7 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.6 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.4 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.3 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.1 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 0.3 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.3 | GO:0023019 | signal transduction involved in regulation of gene expression(GO:0023019) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0071726 | toll-like receptor TLR6:TLR2 signaling pathway(GO:0038124) response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.3 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.5 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |