Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
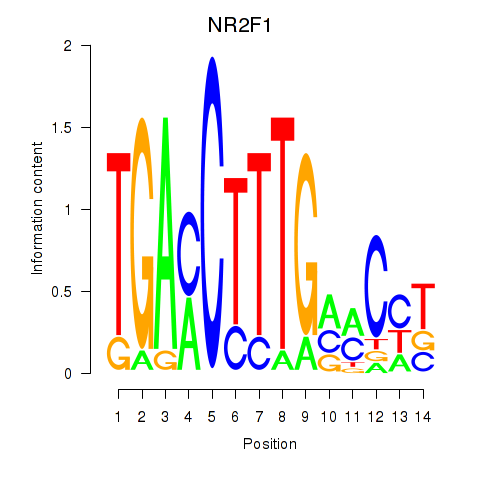
Results for NR2F1
Z-value: 0.49
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | NR2F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F1 | hg19_v2_chr5_+_92919043_92919082 | 0.27 | 3.1e-01 | Click! |
Activity profile of NR2F1 motif
Sorted Z-values of NR2F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_61658853 | 1.05 |
ENST00000525588.1 ENST00000540820.1 |
FADS3 |
fatty acid desaturase 3 |
| chr11_-_61659006 | 0.82 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chr9_-_35685452 | 0.79 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr6_+_143999072 | 0.68 |
ENST00000440869.2 ENST00000367582.3 ENST00000451827.2 |
PHACTR2 |
phosphatase and actin regulator 2 |
| chr16_+_56685796 | 0.64 |
ENST00000334346.2 ENST00000562399.1 |
MT1B |
metallothionein 1B |
| chr10_-_33625154 | 0.60 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr6_-_31926629 | 0.57 |
ENST00000375425.5 ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE |
negative elongation factor complex member E |
| chr7_+_150264365 | 0.57 |
ENST00000255945.2 ENST00000461940.1 |
GIMAP4 |
GTPase, IMAP family member 4 |
| chr5_-_127873659 | 0.53 |
ENST00000262464.4 |
FBN2 |
fibrillin 2 |
| chr3_+_113616317 | 0.50 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chrX_+_66764375 | 0.47 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr20_+_43160409 | 0.43 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr1_-_193075180 | 0.38 |
ENST00000367440.3 |
GLRX2 |
glutaredoxin 2 |
| chrX_-_114253536 | 0.37 |
ENST00000371936.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chr1_+_155294342 | 0.37 |
ENST00000292254.4 |
RUSC1 |
RUN and SH3 domain containing 1 |
| chr2_-_222436988 | 0.37 |
ENST00000409854.1 ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4 |
EPH receptor A4 |
| chr1_+_155294264 | 0.36 |
ENST00000368349.4 |
RUSC1 |
RUN and SH3 domain containing 1 |
| chr12_-_7125770 | 0.36 |
ENST00000261407.4 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
| chr2_+_138721850 | 0.36 |
ENST00000329366.4 ENST00000280097.3 |
HNMT |
histamine N-methyltransferase |
| chr8_+_22436635 | 0.35 |
ENST00000452226.1 ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr11_+_66624527 | 0.35 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr20_+_53092123 | 0.33 |
ENST00000262593.5 |
DOK5 |
docking protein 5 |
| chr20_+_43160458 | 0.32 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_-_111794446 | 0.32 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr11_-_85780853 | 0.29 |
ENST00000531930.1 ENST00000528398.1 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr3_+_113465866 | 0.29 |
ENST00000273398.3 ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr19_-_39390440 | 0.28 |
ENST00000249396.7 ENST00000414941.1 ENST00000392081.2 |
SIRT2 |
sirtuin 2 |
| chr8_+_70404996 | 0.28 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr10_+_123922941 | 0.27 |
ENST00000360561.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr3_-_194393206 | 0.26 |
ENST00000265245.5 |
LSG1 |
large 60S subunit nuclear export GTPase 1 |
| chr16_+_222846 | 0.26 |
ENST00000251595.6 ENST00000397806.1 |
HBA2 |
hemoglobin, alpha 2 |
| chr12_-_51420128 | 0.26 |
ENST00000262051.7 ENST00000547732.1 ENST00000262052.5 ENST00000546488.1 ENST00000550714.1 ENST00000548193.1 ENST00000547579.1 ENST00000546743.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr19_+_45394477 | 0.25 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr19_+_49713991 | 0.25 |
ENST00000597316.1 |
TRPM4 |
transient receptor potential cation channel, subfamily M, member 4 |
| chr4_+_159593418 | 0.25 |
ENST00000507475.1 ENST00000307738.5 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr10_+_104503727 | 0.25 |
ENST00000448841.1 |
WBP1L |
WW domain binding protein 1-like |
| chr7_+_65552756 | 0.24 |
ENST00000450043.1 |
AC068533.7 |
AC068533.7 |
| chr12_-_14996355 | 0.23 |
ENST00000228936.4 |
ART4 |
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr5_+_1801503 | 0.23 |
ENST00000274137.5 ENST00000469176.1 |
NDUFS6 |
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr19_+_39390587 | 0.23 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr12_-_6665200 | 0.23 |
ENST00000336604.4 ENST00000396840.2 ENST00000356896.4 |
IFFO1 |
intermediate filament family orphan 1 |
| chr1_+_38478378 | 0.23 |
ENST00000373014.4 |
UTP11L |
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr8_-_144886321 | 0.23 |
ENST00000526832.1 |
SCRIB |
scribbled planar cell polarity protein |
| chr7_-_115608304 | 0.22 |
ENST00000457268.1 |
TFEC |
transcription factor EC |
| chr1_-_203144941 | 0.21 |
ENST00000255416.4 |
MYBPH |
myosin binding protein H |
| chr1_+_38478432 | 0.21 |
ENST00000537711.1 |
UTP11L |
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr22_+_19950060 | 0.21 |
ENST00000449653.1 |
COMT |
catechol-O-methyltransferase |
| chr1_-_43855479 | 0.21 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr6_-_30080863 | 0.21 |
ENST00000540829.1 |
TRIM31 |
tripartite motif containing 31 |
| chr22_-_19466732 | 0.21 |
ENST00000263202.10 ENST00000360834.4 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr11_-_6640585 | 0.20 |
ENST00000533371.1 ENST00000528657.1 ENST00000436873.2 ENST00000299427.6 |
TPP1 |
tripeptidyl peptidase I |
| chr6_-_30080876 | 0.20 |
ENST00000376734.3 |
TRIM31 |
tripartite motif containing 31 |
| chr12_+_132413765 | 0.20 |
ENST00000376649.3 ENST00000322060.5 |
PUS1 |
pseudouridylate synthase 1 |
| chr1_+_171060018 | 0.20 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr1_-_114302086 | 0.19 |
ENST00000369604.1 ENST00000357783.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr19_+_39616410 | 0.19 |
ENST00000602004.1 ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr1_-_43855444 | 0.19 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr17_-_7307358 | 0.19 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_+_10442993 | 0.19 |
ENST00000423674.1 ENST00000307845.3 |
HPCAL1 |
hippocalcin-like 1 |
| chr2_+_159313452 | 0.18 |
ENST00000389757.3 ENST00000389759.3 |
PKP4 |
plakophilin 4 |
| chr16_+_57673430 | 0.18 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr7_+_120628731 | 0.18 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr8_-_30670053 | 0.17 |
ENST00000518564.1 |
PPP2CB |
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr12_+_49717019 | 0.17 |
ENST00000549275.1 ENST00000551245.1 ENST00000380327.5 ENST00000548311.1 ENST00000550346.1 ENST00000550709.1 ENST00000549534.1 ENST00000257909.3 |
TROAP |
trophinin associated protein |
| chr1_-_114301960 | 0.17 |
ENST00000369598.1 ENST00000369600.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr3_+_9691117 | 0.17 |
ENST00000353332.5 ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14 |
myotubularin related protein 14 |
| chr3_+_37284824 | 0.17 |
ENST00000431105.1 |
GOLGA4 |
golgin A4 |
| chr19_-_51472031 | 0.17 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr17_-_34344991 | 0.17 |
ENST00000591423.1 |
CCL23 |
chemokine (C-C motif) ligand 23 |
| chr14_-_21492251 | 0.16 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr4_-_681114 | 0.16 |
ENST00000503156.1 |
MFSD7 |
major facilitator superfamily domain containing 7 |
| chr12_-_51420108 | 0.15 |
ENST00000547198.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr12_+_132413739 | 0.15 |
ENST00000443358.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr2_-_70475730 | 0.15 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr9_+_139873264 | 0.15 |
ENST00000446677.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr7_-_15601595 | 0.14 |
ENST00000342526.3 |
AGMO |
alkylglycerol monooxygenase |
| chr1_-_114301755 | 0.14 |
ENST00000393357.2 ENST00000369596.2 ENST00000446739.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr12_-_51419924 | 0.14 |
ENST00000541174.2 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr12_+_6494285 | 0.14 |
ENST00000541102.1 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr3_+_128444994 | 0.14 |
ENST00000482525.1 |
RAB7A |
RAB7A, member RAS oncogene family |
| chr11_+_67798090 | 0.14 |
ENST00000313468.5 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr19_-_8408139 | 0.14 |
ENST00000330915.3 ENST00000593649.1 ENST00000595639.1 |
KANK3 |
KN motif and ankyrin repeat domains 3 |
| chr14_-_21492113 | 0.14 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr10_-_75634219 | 0.13 |
ENST00000305762.7 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
| chr1_+_145301735 | 0.13 |
ENST00000605176.1 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr22_+_31518938 | 0.13 |
ENST00000412985.1 ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr7_-_142232071 | 0.13 |
ENST00000390364.3 |
TRBV10-1 |
T cell receptor beta variable 10-1(gene/pseudogene) |
| chr11_-_111957451 | 0.13 |
ENST00000504148.2 ENST00000541231.1 |
TIMM8B |
translocase of inner mitochondrial membrane 8 homolog B (yeast) |
| chr1_-_197036364 | 0.12 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chrX_-_43741594 | 0.12 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr19_+_17420340 | 0.12 |
ENST00000359866.4 |
DDA1 |
DET1 and DDB1 associated 1 |
| chr2_+_44502597 | 0.12 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr11_-_26743546 | 0.12 |
ENST00000280467.6 ENST00000396005.3 |
SLC5A12 |
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr11_+_64073699 | 0.12 |
ENST00000405666.1 ENST00000468670.1 |
ESRRA |
estrogen-related receptor alpha |
| chr16_+_67207838 | 0.12 |
ENST00000566871.1 ENST00000268605.7 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr19_-_12780211 | 0.12 |
ENST00000597961.1 ENST00000598732.1 ENST00000222190.5 |
CTD-2192J16.24 WDR83OS |
Uncharacterized protein WD repeat domain 83 opposite strand |
| chr19_-_51472222 | 0.12 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr17_-_34345002 | 0.12 |
ENST00000293280.2 |
CCL23 |
chemokine (C-C motif) ligand 23 |
| chr1_-_114301503 | 0.12 |
ENST00000447664.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr11_+_67798114 | 0.12 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr11_-_124767693 | 0.11 |
ENST00000533054.1 |
ROBO4 |
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr20_-_23860373 | 0.11 |
ENST00000304710.4 |
CST5 |
cystatin D |
| chr9_-_27005686 | 0.11 |
ENST00000380055.5 |
LRRC19 |
leucine rich repeat containing 19 |
| chr2_-_165698521 | 0.11 |
ENST00000409184.3 ENST00000392717.2 ENST00000456693.1 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr11_+_124543694 | 0.11 |
ENST00000227135.2 ENST00000532692.1 |
SPA17 |
sperm autoantigenic protein 17 |
| chr6_-_32145861 | 0.11 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr3_+_37284668 | 0.11 |
ENST00000361924.2 ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4 |
golgin A4 |
| chr10_-_17171817 | 0.11 |
ENST00000377833.4 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
| chr16_+_67207872 | 0.11 |
ENST00000563258.1 ENST00000568146.1 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr9_+_34458771 | 0.10 |
ENST00000437363.1 ENST00000242317.4 |
DNAI1 |
dynein, axonemal, intermediate chain 1 |
| chr22_-_24641027 | 0.10 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr1_+_176432298 | 0.10 |
ENST00000367661.3 ENST00000367662.3 |
PAPPA2 |
pappalysin 2 |
| chr3_+_128444965 | 0.10 |
ENST00000265062.3 |
RAB7A |
RAB7A, member RAS oncogene family |
| chr2_-_70475701 | 0.10 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr2_+_220491973 | 0.09 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr19_+_10222189 | 0.09 |
ENST00000321826.4 |
P2RY11 |
purinergic receptor P2Y, G-protein coupled, 11 |
| chr12_-_6960407 | 0.09 |
ENST00000540683.1 ENST00000229265.6 ENST00000535406.1 ENST00000422785.3 |
CDCA3 |
cell division cycle associated 3 |
| chr10_-_96829246 | 0.09 |
ENST00000371270.3 ENST00000535898.1 ENST00000539050.1 |
CYP2C8 |
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr5_+_140593509 | 0.09 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chr3_-_113465065 | 0.09 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr12_+_132413798 | 0.09 |
ENST00000440818.2 ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr4_+_159593271 | 0.09 |
ENST00000512251.1 ENST00000511912.1 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr2_-_219134343 | 0.09 |
ENST00000447885.1 ENST00000420660.1 |
AAMP |
angio-associated, migratory cell protein |
| chr4_-_129209944 | 0.09 |
ENST00000520121.1 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr15_-_73925651 | 0.09 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr7_-_107204918 | 0.09 |
ENST00000297135.3 |
COG5 |
component of oligomeric golgi complex 5 |
| chr11_-_66445219 | 0.09 |
ENST00000525754.1 ENST00000531969.1 ENST00000524637.1 ENST00000531036.2 ENST00000310046.4 |
RBM4B |
RNA binding motif protein 4B |
| chr1_+_156084461 | 0.08 |
ENST00000347559.2 ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA |
lamin A/C |
| chr10_+_18689637 | 0.08 |
ENST00000377315.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr3_+_10290596 | 0.08 |
ENST00000448281.2 |
TATDN2 |
TatD DNase domain containing 2 |
| chr1_-_109618566 | 0.08 |
ENST00000338366.5 |
TAF13 |
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr11_+_113848216 | 0.08 |
ENST00000299961.5 |
HTR3A |
5-hydroxytryptamine (serotonin) receptor 3A, ionotropic |
| chr10_-_74114714 | 0.08 |
ENST00000338820.3 ENST00000394903.2 ENST00000444643.2 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr1_-_161102421 | 0.08 |
ENST00000490843.2 ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD |
death effector domain containing |
| chr14_+_101299520 | 0.08 |
ENST00000455531.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr2_-_178128528 | 0.08 |
ENST00000397063.4 ENST00000421929.1 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr16_+_81272287 | 0.08 |
ENST00000425577.2 ENST00000564552.1 |
BCMO1 |
beta-carotene 15,15'-monooxygenase 1 |
| chr1_+_158149737 | 0.07 |
ENST00000368171.3 |
CD1D |
CD1d molecule |
| chr19_+_11546093 | 0.07 |
ENST00000591462.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr1_+_109234907 | 0.07 |
ENST00000370025.4 ENST00000370022.5 ENST00000370021.1 |
PRPF38B |
pre-mRNA processing factor 38B |
| chr15_+_58724184 | 0.07 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr6_+_24775153 | 0.07 |
ENST00000356509.3 ENST00000230056.3 |
GMNN |
geminin, DNA replication inhibitor |
| chr5_-_148930960 | 0.07 |
ENST00000261798.5 ENST00000377843.2 |
CSNK1A1 |
casein kinase 1, alpha 1 |
| chr16_-_4292071 | 0.07 |
ENST00000399609.3 |
SRL |
sarcalumenin |
| chr17_+_42264395 | 0.06 |
ENST00000587989.1 ENST00000590235.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr20_-_7921090 | 0.06 |
ENST00000378789.3 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr9_+_91933407 | 0.06 |
ENST00000375807.3 ENST00000339901.4 |
SECISBP2 |
SECIS binding protein 2 |
| chrX_+_101478829 | 0.06 |
ENST00000372763.1 ENST00000372758.1 |
NXF2 |
nuclear RNA export factor 2 |
| chr2_-_152382500 | 0.06 |
ENST00000434685.1 |
NEB |
nebulin |
| chr1_+_196946680 | 0.06 |
ENST00000256785.4 |
CFHR5 |
complement factor H-related 5 |
| chr20_-_44485835 | 0.06 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr1_-_151299842 | 0.06 |
ENST00000438243.2 ENST00000489223.2 ENST00000368873.1 ENST00000430800.1 ENST00000368872.1 |
PI4KB |
phosphatidylinositol 4-kinase, catalytic, beta |
| chr11_+_72929319 | 0.06 |
ENST00000393597.2 ENST00000311131.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr20_+_54987168 | 0.06 |
ENST00000360314.3 |
CASS4 |
Cas scaffolding protein family member 4 |
| chr11_-_73720276 | 0.06 |
ENST00000348534.4 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr1_+_43855560 | 0.06 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr1_-_6420737 | 0.06 |
ENST00000541130.1 ENST00000377845.3 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chr7_-_37956409 | 0.05 |
ENST00000436072.2 |
SFRP4 |
secreted frizzled-related protein 4 |
| chr7_-_75452673 | 0.05 |
ENST00000416943.1 |
CCL24 |
chemokine (C-C motif) ligand 24 |
| chr7_-_74489609 | 0.05 |
ENST00000329959.4 ENST00000503250.2 ENST00000543840.1 |
WBSCR16 |
Williams-Beuren syndrome chromosome region 16 |
| chr2_-_70780572 | 0.05 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr6_+_31926857 | 0.05 |
ENST00000375394.2 ENST00000544581.1 |
SKIV2L |
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr18_+_54318616 | 0.05 |
ENST00000254442.3 |
WDR7 |
WD repeat domain 7 |
| chr1_-_211848899 | 0.05 |
ENST00000366998.3 ENST00000540251.1 ENST00000366999.4 |
NEK2 |
NIMA-related kinase 2 |
| chr15_+_40453204 | 0.05 |
ENST00000287598.6 ENST00000412359.3 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr19_+_50919056 | 0.05 |
ENST00000599632.1 |
CTD-2545M3.6 |
CTD-2545M3.6 |
| chr17_-_4463856 | 0.05 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr18_+_54318566 | 0.05 |
ENST00000589935.1 ENST00000357574.3 |
WDR7 |
WD repeat domain 7 |
| chr2_-_38978492 | 0.05 |
ENST00000409276.1 ENST00000446327.2 ENST00000313117.6 |
SRSF7 |
serine/arginine-rich splicing factor 7 |
| chr2_-_219134822 | 0.05 |
ENST00000444053.1 ENST00000248450.4 |
AAMP |
angio-associated, migratory cell protein |
| chr11_+_67798363 | 0.05 |
ENST00000525628.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr20_+_54987305 | 0.05 |
ENST00000371336.3 ENST00000434344.1 |
CASS4 |
Cas scaffolding protein family member 4 |
| chrX_-_108868390 | 0.05 |
ENST00000372101.2 |
KCNE1L |
KCNE1-like |
| chr1_+_196946664 | 0.05 |
ENST00000367414.5 |
CFHR5 |
complement factor H-related 5 |
| chr11_+_72929402 | 0.04 |
ENST00000393596.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr1_-_109968973 | 0.04 |
ENST00000271308.4 ENST00000538610.1 |
PSMA5 |
proteasome (prosome, macropain) subunit, alpha type, 5 |
| chr1_-_161102367 | 0.04 |
ENST00000464113.1 |
DEDD |
death effector domain containing |
| chr4_-_11431188 | 0.04 |
ENST00000510712.1 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr11_-_73720122 | 0.04 |
ENST00000426995.2 |
UCP3 |
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr1_-_151735937 | 0.04 |
ENST00000368829.3 ENST00000368830.3 |
MRPL9 |
mitochondrial ribosomal protein L9 |
| chr14_-_39572345 | 0.04 |
ENST00000548032.2 ENST00000556092.1 ENST00000557280.1 ENST00000545328.2 ENST00000553970.1 |
SEC23A |
Sec23 homolog A (S. cerevisiae) |
| chr1_+_156785425 | 0.04 |
ENST00000392302.2 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
| chr19_+_535835 | 0.04 |
ENST00000607527.1 ENST00000606065.1 |
CDC34 |
cell division cycle 34 |
| chr6_+_32146131 | 0.04 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr6_+_97010424 | 0.04 |
ENST00000541107.1 ENST00000450218.1 ENST00000326771.2 |
FHL5 |
four and a half LIM domains 5 |
| chr8_-_80942139 | 0.04 |
ENST00000521434.1 ENST00000519120.1 ENST00000520946.1 |
MRPS28 |
mitochondrial ribosomal protein S28 |
| chr6_+_30594619 | 0.04 |
ENST00000318999.7 ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
| chr19_-_10687907 | 0.03 |
ENST00000589348.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr7_-_38948774 | 0.03 |
ENST00000395969.2 ENST00000414632.1 ENST00000310301.4 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr1_-_155270770 | 0.03 |
ENST00000392414.3 |
PKLR |
pyruvate kinase, liver and RBC |
| chr3_-_49449350 | 0.03 |
ENST00000454011.2 ENST00000445425.1 ENST00000422781.1 |
RHOA |
ras homolog family member A |
| chrX_+_138612889 | 0.03 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr10_+_26986582 | 0.03 |
ENST00000376215.5 ENST00000376203.5 |
PDSS1 |
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr19_+_44100727 | 0.03 |
ENST00000528387.1 ENST00000529930.1 ENST00000336564.4 ENST00000607544.1 ENST00000526798.1 |
ZNF576 SRRM5 |
zinc finger protein 576 serine/arginine repetitive matrix 5 |
| chr1_-_36235559 | 0.03 |
ENST00000251195.5 |
CLSPN |
claspin |
| chr16_+_3019552 | 0.03 |
ENST00000572687.1 |
PAQR4 |
progestin and adipoQ receptor family member IV |
| chr11_+_63137251 | 0.03 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr19_+_44100632 | 0.03 |
ENST00000533118.1 |
ZNF576 |
zinc finger protein 576 |
| chr2_-_28113217 | 0.03 |
ENST00000444339.2 |
RBKS |
ribokinase |
| chr19_+_11546153 | 0.03 |
ENST00000591946.1 ENST00000252455.2 ENST00000412601.1 |
PRKCSH |
protein kinase C substrate 80K-H |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.6 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.6 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.0 | 0.3 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.7 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.3 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0034706 | sodium channel complex(GO:0034706) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 0.6 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.3 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.4 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.1 | 0.4 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.6 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.5 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.1 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.3 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.1 | 0.4 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 0.4 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 0.6 | GO:0060301 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.1 | 0.6 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.3 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.2 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.1 | 0.2 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.3 | GO:1904199 | positive regulation of cardiac conduction(GO:1903781) positive regulation of atrial cardiac muscle cell action potential(GO:1903949) positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.5 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) positive regulation of sulfur amino acid metabolic process(GO:0031337) positive regulation of homocysteine metabolic process(GO:0050668) |
| 0.0 | 0.4 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.3 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.6 | GO:0034244 | negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.3 | GO:0060686 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.4 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 1.9 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.3 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.3 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0046223 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.8 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.3 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.0 | GO:0043366 | beta selection(GO:0043366) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0009441 | glycolate metabolic process(GO:0009441) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |