Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
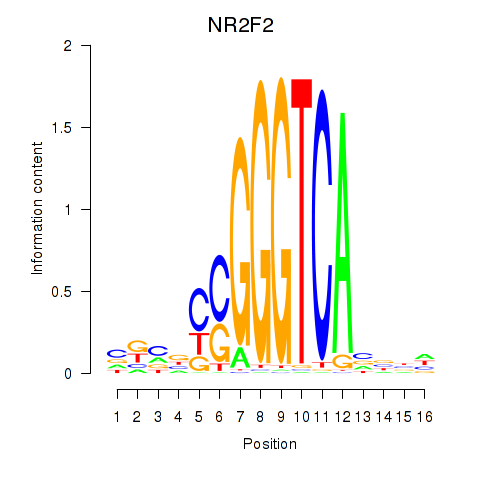
Results for NR2F2
Z-value: 0.71
Transcription factors associated with NR2F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F2
|
ENSG00000185551.8 | NR2F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F2 | hg19_v2_chr15_+_96876340_96876392 | 0.83 | 6.8e-05 | Click! |
Activity profile of NR2F2 motif
Sorted Z-values of NR2F2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_33010175 | 1.28 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr10_+_89419370 | 1.19 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_-_151344172 | 1.16 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr2_+_189157536 | 1.13 |
ENST00000409580.1 ENST00000409637.3 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_189157498 | 1.13 |
ENST00000359135.3 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr2_+_192110199 | 1.02 |
ENST00000304164.4 |
MYO1B |
myosin IB |
| chr11_+_12132117 | 0.97 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr7_+_79764104 | 0.94 |
ENST00000351004.3 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr7_-_107642348 | 0.93 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chr1_-_156647189 | 0.79 |
ENST00000368223.3 |
NES |
nestin |
| chr10_-_15413035 | 0.78 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chr1_-_20812690 | 0.78 |
ENST00000375078.3 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
| chr20_+_43160409 | 0.77 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr6_+_142623063 | 0.73 |
ENST00000296932.8 ENST00000367609.3 |
GPR126 |
G protein-coupled receptor 126 |
| chr6_-_2971792 | 0.71 |
ENST00000380546.3 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr6_-_2971429 | 0.70 |
ENST00000380529.1 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr9_+_124030338 | 0.69 |
ENST00000449773.1 ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN |
gelsolin |
| chr4_-_99579733 | 0.68 |
ENST00000305798.3 |
TSPAN5 |
tetraspanin 5 |
| chr2_-_175870085 | 0.68 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr7_-_45960850 | 0.67 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chr17_-_1619491 | 0.67 |
ENST00000570416.1 ENST00000575626.1 ENST00000610106.1 ENST00000608198.1 ENST00000609442.1 ENST00000334146.3 ENST00000576489.1 ENST00000608245.1 ENST00000609398.1 ENST00000608913.1 ENST00000574016.1 ENST00000571091.1 ENST00000573127.1 ENST00000609990.1 ENST00000576749.1 |
MIR22HG |
MIR22 host gene (non-protein coding) |
| chr21_+_47518011 | 0.66 |
ENST00000300527.4 ENST00000357838.4 ENST00000310645.5 |
COL6A2 |
collagen, type VI, alpha 2 |
| chr14_-_94856987 | 0.65 |
ENST00000449399.3 ENST00000404814.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr14_-_94857004 | 0.63 |
ENST00000557492.1 ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr14_-_94856951 | 0.63 |
ENST00000553327.1 ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr6_-_2971494 | 0.62 |
ENST00000380539.1 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
| chr13_+_113951532 | 0.61 |
ENST00000332556.4 |
LAMP1 |
lysosomal-associated membrane protein 1 |
| chr3_+_45067659 | 0.60 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr19_-_43702231 | 0.60 |
ENST00000597374.1 ENST00000599371.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chrX_+_99899180 | 0.58 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr18_+_19749386 | 0.57 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr19_-_6720686 | 0.55 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr16_+_66400533 | 0.54 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr2_+_85811525 | 0.54 |
ENST00000306384.4 |
VAMP5 |
vesicle-associated membrane protein 5 |
| chr5_+_66124590 | 0.53 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr13_-_40177261 | 0.53 |
ENST00000379589.3 |
LHFP |
lipoma HMGIC fusion partner |
| chr7_+_116139744 | 0.52 |
ENST00000343213.2 |
CAV2 |
caveolin 2 |
| chr1_-_57045228 | 0.51 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr2_+_102928009 | 0.51 |
ENST00000404917.2 ENST00000447231.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr22_-_19512893 | 0.50 |
ENST00000403084.1 ENST00000413119.2 |
CLDN5 |
claudin 5 |
| chr11_-_27722021 | 0.50 |
ENST00000356660.4 ENST00000418212.1 ENST00000533246.1 |
BDNF |
brain-derived neurotrophic factor |
| chrX_+_100333709 | 0.49 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr2_-_3523507 | 0.49 |
ENST00000327435.6 |
ADI1 |
acireductone dioxygenase 1 |
| chr5_-_131563501 | 0.49 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr20_+_44034804 | 0.48 |
ENST00000357275.2 ENST00000372720.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr2_+_201450591 | 0.48 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr14_+_61447927 | 0.48 |
ENST00000451406.1 |
SLC38A6 |
solute carrier family 38, member 6 |
| chr11_-_61658853 | 0.48 |
ENST00000525588.1 ENST00000540820.1 |
FADS3 |
fatty acid desaturase 3 |
| chr14_+_61447832 | 0.47 |
ENST00000354886.2 ENST00000267488.4 |
SLC38A6 |
solute carrier family 38, member 6 |
| chr5_-_131562935 | 0.47 |
ENST00000379104.2 ENST00000379100.2 ENST00000428369.1 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr6_-_2842087 | 0.47 |
ENST00000537185.1 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chrX_+_54834791 | 0.47 |
ENST00000218439.4 ENST00000375058.1 ENST00000375060.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr12_+_102091400 | 0.46 |
ENST00000229266.3 ENST00000549872.1 |
CHPT1 |
choline phosphotransferase 1 |
| chr1_+_43996518 | 0.46 |
ENST00000359947.4 ENST00000438120.1 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
| chr1_+_223889285 | 0.46 |
ENST00000433674.2 |
CAPN2 |
calpain 2, (m/II) large subunit |
| chr14_-_74959978 | 0.46 |
ENST00000541064.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr14_-_74960030 | 0.45 |
ENST00000553490.1 ENST00000557510.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chrX_+_54834004 | 0.44 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr22_+_35776828 | 0.44 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr9_-_116840728 | 0.43 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr14_-_74959994 | 0.43 |
ENST00000238633.2 ENST00000434013.2 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr6_-_2842219 | 0.43 |
ENST00000380739.5 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr6_+_53659746 | 0.43 |
ENST00000370888.1 |
LRRC1 |
leucine rich repeat containing 1 |
| chr20_+_43343886 | 0.42 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr2_+_236402669 | 0.42 |
ENST00000409457.1 ENST00000336665.5 ENST00000304032.8 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr20_+_44034676 | 0.40 |
ENST00000372723.3 ENST00000372722.3 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_+_54694119 | 0.40 |
ENST00000456872.1 ENST00000302937.4 ENST00000429671.2 |
TSEN34 |
TSEN34 tRNA splicing endonuclease subunit |
| chr19_-_43269809 | 0.39 |
ENST00000406636.3 ENST00000404209.4 ENST00000306511.4 |
PSG8 |
pregnancy specific beta-1-glycoprotein 8 |
| chr9_-_79009414 | 0.39 |
ENST00000376736.1 |
RFK |
riboflavin kinase |
| chr17_-_41174424 | 0.38 |
ENST00000355653.3 |
VAT1 |
vesicle amine transport 1 |
| chr11_-_61659006 | 0.37 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chr9_-_79009048 | 0.37 |
ENST00000490113.1 |
RFK |
riboflavin kinase |
| chr10_-_90751038 | 0.37 |
ENST00000458159.1 ENST00000415557.1 ENST00000458208.1 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr19_-_54693401 | 0.36 |
ENST00000338624.6 |
MBOAT7 |
membrane bound O-acyltransferase domain containing 7 |
| chrX_+_54834159 | 0.36 |
ENST00000375053.2 ENST00000347546.4 ENST00000375062.4 |
MAGED2 |
melanoma antigen family D, 2 |
| chr12_+_52626898 | 0.35 |
ENST00000331817.5 |
KRT7 |
keratin 7 |
| chr17_+_41006095 | 0.35 |
ENST00000591562.1 ENST00000588033.1 |
AOC3 |
amine oxidase, copper containing 3 |
| chr20_+_44035200 | 0.35 |
ENST00000372717.1 ENST00000360981.4 |
DBNDD2 |
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr12_+_6493199 | 0.35 |
ENST00000228918.4 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chrX_-_43741594 | 0.34 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr20_+_44519948 | 0.34 |
ENST00000354880.5 ENST00000191018.5 |
CTSA |
cathepsin A |
| chr19_-_42463418 | 0.32 |
ENST00000600292.1 ENST00000601078.1 ENST00000601891.1 ENST00000222008.6 |
RABAC1 |
Rab acceptor 1 (prenylated) |
| chr20_+_44520009 | 0.31 |
ENST00000607482.1 ENST00000372459.2 |
CTSA |
cathepsin A |
| chr17_+_78075361 | 0.31 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chrX_+_54835493 | 0.31 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr7_+_143079000 | 0.31 |
ENST00000392910.2 |
ZYX |
zyxin |
| chr17_+_78075498 | 0.31 |
ENST00000302262.3 |
GAA |
glucosidase, alpha; acid |
| chr16_-_53537105 | 0.31 |
ENST00000568596.1 ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP |
AKT interacting protein |
| chr7_+_94139105 | 0.31 |
ENST00000297273.4 |
CASD1 |
CAS1 domain containing 1 |
| chr6_-_105627735 | 0.31 |
ENST00000254765.3 |
POPDC3 |
popeye domain containing 3 |
| chr11_-_33795893 | 0.30 |
ENST00000526785.1 ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3 |
F-box protein 3 |
| chr3_-_160823040 | 0.30 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr17_-_80291818 | 0.30 |
ENST00000269389.3 ENST00000581691.1 |
SECTM1 |
secreted and transmembrane 1 |
| chr2_+_201170770 | 0.30 |
ENST00000409988.3 ENST00000409385.1 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr11_-_65308082 | 0.29 |
ENST00000532661.1 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr1_-_95007193 | 0.29 |
ENST00000370207.4 ENST00000334047.7 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
| chr9_+_116263639 | 0.29 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr1_-_33168336 | 0.29 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr2_-_235405679 | 0.29 |
ENST00000390645.2 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr18_+_32556892 | 0.28 |
ENST00000591734.1 ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr22_+_44319648 | 0.28 |
ENST00000423180.2 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr1_-_236228403 | 0.28 |
ENST00000366595.3 |
NID1 |
nidogen 1 |
| chr13_-_52733510 | 0.28 |
ENST00000378101.2 |
NEK3 |
NIMA-related kinase 3 |
| chr21_-_46707793 | 0.28 |
ENST00000331343.7 ENST00000349485.5 |
POFUT2 |
protein O-fucosyltransferase 2 |
| chr3_-_45883558 | 0.28 |
ENST00000445698.1 ENST00000296135.6 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
| chr22_+_44319619 | 0.28 |
ENST00000216180.3 |
PNPLA3 |
patatin-like phospholipase domain containing 3 |
| chr12_+_102271129 | 0.27 |
ENST00000258534.8 |
DRAM1 |
DNA-damage regulated autophagy modulator 1 |
| chr7_+_143078652 | 0.27 |
ENST00000354434.4 ENST00000449423.2 |
ZYX |
zyxin |
| chr15_-_63448973 | 0.27 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr2_-_86790593 | 0.27 |
ENST00000263856.4 ENST00000409225.2 |
CHMP3 |
charged multivesicular body protein 3 |
| chr20_+_8112824 | 0.26 |
ENST00000378641.3 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
| chr8_-_145028013 | 0.26 |
ENST00000354958.2 |
PLEC |
plectin |
| chr10_-_118765081 | 0.26 |
ENST00000392903.2 ENST00000355371.4 |
KIAA1598 |
KIAA1598 |
| chr12_+_110011571 | 0.26 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr7_+_120629653 | 0.26 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr7_+_112090483 | 0.25 |
ENST00000403825.3 ENST00000429071.1 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr17_+_79650962 | 0.25 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr11_-_2906979 | 0.25 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr6_-_131321863 | 0.25 |
ENST00000528282.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr21_+_47531328 | 0.25 |
ENST00000409416.1 ENST00000397763.1 |
COL6A2 |
collagen, type VI, alpha 2 |
| chr1_+_207925391 | 0.24 |
ENST00000358170.2 ENST00000354848.1 ENST00000322918.5 ENST00000367042.1 ENST00000367041.1 ENST00000357714.1 ENST00000322875.4 ENST00000367047.1 ENST00000441839.2 ENST00000361067.1 ENST00000360212.2 ENST00000480003.1 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr7_-_151433342 | 0.24 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chrX_+_30671476 | 0.24 |
ENST00000378946.3 ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK |
glycerol kinase |
| chr11_-_89224139 | 0.24 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr21_-_43187231 | 0.23 |
ENST00000332512.3 ENST00000352483.2 |
RIPK4 |
receptor-interacting serine-threonine kinase 4 |
| chr5_+_150827143 | 0.23 |
ENST00000243389.3 ENST00000517945.1 ENST00000521925.1 |
SLC36A1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr15_-_66790146 | 0.23 |
ENST00000316634.5 |
SNAPC5 |
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr2_-_264024 | 0.23 |
ENST00000403712.2 ENST00000356150.5 ENST00000405430.1 |
SH3YL1 |
SH3 and SYLF domain containing 1 |
| chr7_-_151433393 | 0.23 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr6_-_46459099 | 0.23 |
ENST00000371374.1 |
RCAN2 |
regulator of calcineurin 2 |
| chr1_-_43855479 | 0.23 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr15_-_89456630 | 0.23 |
ENST00000268150.8 |
MFGE8 |
milk fat globule-EGF factor 8 protein |
| chr14_-_75536182 | 0.22 |
ENST00000555463.1 |
ACYP1 |
acylphosphatase 1, erythrocyte (common) type |
| chr17_-_61523535 | 0.22 |
ENST00000584031.1 ENST00000392976.1 |
CYB561 |
cytochrome b561 |
| chr6_-_80657292 | 0.21 |
ENST00000369816.4 |
ELOVL4 |
ELOVL fatty acid elongase 4 |
| chr6_+_7541845 | 0.21 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr17_+_19437132 | 0.21 |
ENST00000436810.2 ENST00000270570.4 ENST00000457293.1 ENST00000542886.1 ENST00000575023.1 ENST00000395585.1 |
SLC47A1 |
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr11_-_1587166 | 0.20 |
ENST00000331588.4 |
DUSP8 |
dual specificity phosphatase 8 |
| chr12_-_65153175 | 0.20 |
ENST00000543646.1 ENST00000542058.1 ENST00000258145.3 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
| chr16_-_89785777 | 0.20 |
ENST00000561976.1 |
VPS9D1 |
VPS9 domain containing 1 |
| chr19_-_54693521 | 0.20 |
ENST00000391754.1 ENST00000245615.1 ENST00000431666.2 |
MBOAT7 |
membrane bound O-acyltransferase domain containing 7 |
| chr19_+_17337027 | 0.20 |
ENST00000601529.1 ENST00000600232.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr4_+_8201091 | 0.20 |
ENST00000382521.3 ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1 |
SH3 domain and tetratricopeptide repeats 1 |
| chr3_+_49059038 | 0.20 |
ENST00000451378.2 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr19_+_17337007 | 0.19 |
ENST00000215061.4 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr5_+_68530668 | 0.19 |
ENST00000506563.1 |
CDK7 |
cyclin-dependent kinase 7 |
| chr1_-_48937838 | 0.19 |
ENST00000371847.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr1_-_43855444 | 0.19 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr1_-_173174681 | 0.19 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr1_-_17307173 | 0.19 |
ENST00000438542.1 ENST00000375535.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr16_+_524850 | 0.19 |
ENST00000450428.1 ENST00000452814.1 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
| chr13_-_36920872 | 0.19 |
ENST00000451493.1 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr17_-_7145475 | 0.19 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr1_-_48937821 | 0.19 |
ENST00000396199.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr4_-_85887503 | 0.18 |
ENST00000509172.1 ENST00000322366.6 ENST00000295888.4 ENST00000502713.1 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
| chr13_-_36920615 | 0.18 |
ENST00000494062.2 |
SPG20 |
spastic paraplegia 20 (Troyer syndrome) |
| chr9_-_21335240 | 0.18 |
ENST00000537938.1 |
KLHL9 |
kelch-like family member 9 |
| chr3_-_122512619 | 0.18 |
ENST00000383659.1 ENST00000306103.2 |
HSPBAP1 |
HSPB (heat shock 27kDa) associated protein 1 |
| chrX_+_101975643 | 0.18 |
ENST00000361229.4 |
BHLHB9 |
basic helix-loop-helix domain containing, class B, 9 |
| chr3_-_196695692 | 0.18 |
ENST00000412723.1 |
PIGZ |
phosphatidylinositol glycan anchor biosynthesis, class Z |
| chr19_+_17337406 | 0.18 |
ENST00000597836.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr1_+_87595433 | 0.18 |
ENST00000469312.2 ENST00000490006.2 |
RP5-1052I5.1 |
long intergenic non-protein coding RNA 1140 |
| chr16_-_30134266 | 0.18 |
ENST00000484663.1 ENST00000478356.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr1_+_23037323 | 0.18 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr3_+_42947600 | 0.18 |
ENST00000328199.6 ENST00000541208.1 |
ZNF662 |
zinc finger protein 662 |
| chr5_+_68530697 | 0.18 |
ENST00000256443.3 ENST00000514676.1 |
CDK7 |
cyclin-dependent kinase 7 |
| chrX_+_101975619 | 0.18 |
ENST00000457056.1 |
BHLHB9 |
basic helix-loop-helix domain containing, class B, 9 |
| chr3_-_50329835 | 0.17 |
ENST00000429673.2 |
IFRD2 |
interferon-related developmental regulator 2 |
| chr11_-_89224638 | 0.17 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr10_+_135207598 | 0.17 |
ENST00000477902.2 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr5_-_114961858 | 0.17 |
ENST00000282382.4 ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2 TMED7 TICAM2 |
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr10_-_52645416 | 0.17 |
ENST00000374001.2 ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF |
APOBEC1 complementation factor |
| chr5_+_131593364 | 0.17 |
ENST00000253754.3 ENST00000379018.3 |
PDLIM4 |
PDZ and LIM domain 4 |
| chr6_+_31865552 | 0.17 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr16_-_84150492 | 0.17 |
ENST00000343411.3 |
MBTPS1 |
membrane-bound transcription factor peptidase, site 1 |
| chr1_-_244013384 | 0.17 |
ENST00000366539.1 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr4_-_186697044 | 0.17 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr22_-_19165917 | 0.17 |
ENST00000451283.1 |
SLC25A1 |
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr11_-_65325430 | 0.16 |
ENST00000322147.4 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
| chr14_-_105262016 | 0.16 |
ENST00000407796.2 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr12_+_132434439 | 0.16 |
ENST00000333577.4 ENST00000389561.2 ENST00000389562.2 ENST00000332482.4 ENST00000330386.6 |
EP400 |
E1A binding protein p400 |
| chr11_-_85780086 | 0.16 |
ENST00000532317.1 ENST00000528256.1 ENST00000526033.1 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr1_+_160313062 | 0.16 |
ENST00000294785.5 ENST00000368063.1 ENST00000437169.1 |
NCSTN |
nicastrin |
| chr11_-_85779971 | 0.16 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr10_-_49812997 | 0.16 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr11_-_89224488 | 0.15 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89224299 | 0.15 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr20_-_33413416 | 0.15 |
ENST00000359003.2 |
NCOA6 |
nuclear receptor coactivator 6 |
| chr10_-_52645379 | 0.15 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr9_-_21335356 | 0.15 |
ENST00000359039.4 |
KLHL9 |
kelch-like family member 9 |
| chr8_-_57358432 | 0.15 |
ENST00000517415.1 ENST00000314922.3 |
PENK |
proenkephalin |
| chr17_-_53809473 | 0.15 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr19_-_47987419 | 0.15 |
ENST00000536339.1 ENST00000595554.1 ENST00000600271.1 ENST00000338134.3 |
KPTN |
kaptin (actin binding protein) |
| chr22_+_45898712 | 0.15 |
ENST00000455233.1 ENST00000348697.2 ENST00000402984.3 ENST00000262722.7 ENST00000327858.6 ENST00000442170.2 ENST00000340923.5 ENST00000439835.1 |
FBLN1 |
fibulin 1 |
| chr4_+_159593418 | 0.15 |
ENST00000507475.1 ENST00000307738.5 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr22_+_38302285 | 0.15 |
ENST00000215957.6 |
MICALL1 |
MICAL-like 1 |
| chr14_-_105262055 | 0.15 |
ENST00000349310.3 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr12_+_113659234 | 0.15 |
ENST00000551096.1 ENST00000551099.1 ENST00000335509.6 ENST00000552897.1 ENST00000550785.1 ENST00000549279.1 |
TPCN1 |
two pore segment channel 1 |
| chr16_+_57673430 | 0.15 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr13_+_113951607 | 0.15 |
ENST00000397181.3 |
LAMP1 |
lysosomal-associated membrane protein 1 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 6.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.3 | 1.3 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.3 | 1.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 1.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.3 | 0.8 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.2 | 0.9 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.2 | 1.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.2 | 0.6 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 0.8 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 | 0.5 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.5 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 0.5 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.2 | 0.6 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) |
| 0.1 | 0.7 | GO:0044858 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 0.5 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.5 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.1 | 0.3 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.1 | 0.6 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.1 | 0.3 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.2 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.1 | 0.3 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.1 | 0.5 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 1.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.5 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.1 | 0.3 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.5 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.4 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.7 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.1 | 0.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.1 | 1.0 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.3 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.1 | 0.1 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 0.1 | 0.4 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.1 | 0.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.1 | 0.6 | GO:0036155 | acylglycerol acyl-chain remodeling(GO:0036155) |
| 0.1 | 0.3 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.5 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.8 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.9 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.3 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.1 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.1 | GO:1901253 | negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.0 | 0.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.6 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.3 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.4 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.2 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.3 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) |
| 0.0 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.0 | 0.4 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.7 | GO:0044342 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.7 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 2.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.6 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.3 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 1.4 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.2 | GO:0005975 | carbohydrate metabolic process(GO:0005975) |
| 0.0 | 0.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.7 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.0 | 0.1 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.4 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 | 0.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.3 | GO:1903541 | regulation of exosomal secretion(GO:1903541) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.3 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.0 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 0.0 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.0 | GO:0050720 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) interleukin-1 beta biosynthetic process(GO:0050720) |
| 0.0 | 0.1 | GO:1903147 | negative regulation of macromitophagy(GO:1901525) negative regulation of mitophagy(GO:1903147) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.8 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.6 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 2.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.6 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 0.6 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.2 | 1.0 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.3 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 0.5 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.4 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.5 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.4 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 0.5 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.7 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.4 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.7 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.4 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.7 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 0.7 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.6 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0000995 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.3 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.3 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.5 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.0 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.8 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.9 | GO:0001618 | virus receptor activity(GO:0001618) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.5 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.3 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.1 | 0.5 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.8 | GO:0032010 | phagolysosome(GO:0032010) alveolar lamellar body(GO:0097208) |
| 0.1 | 0.7 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.3 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.1 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 2.8 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 3.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 2.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.5 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 1.0 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.0 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |