Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for NR3C1
Z-value: 1.51
Transcription factors associated with NR3C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR3C1
|
ENSG00000113580.10 | NR3C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR3C1 | hg19_v2_chr5_-_142814241_142814278, hg19_v2_chr5_-_142783694_142783842 | 0.72 | 1.8e-03 | Click! |
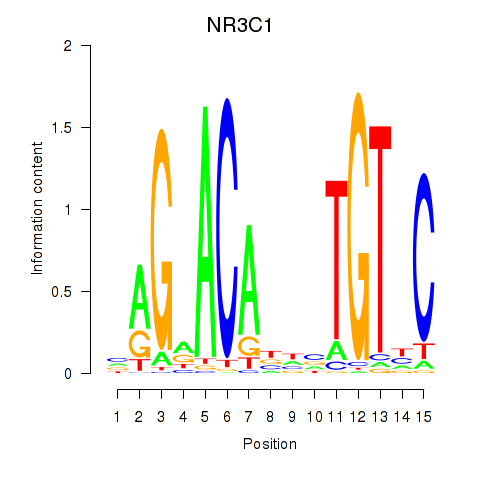
Activity profile of NR3C1 motif
Sorted Z-values of NR3C1 motif
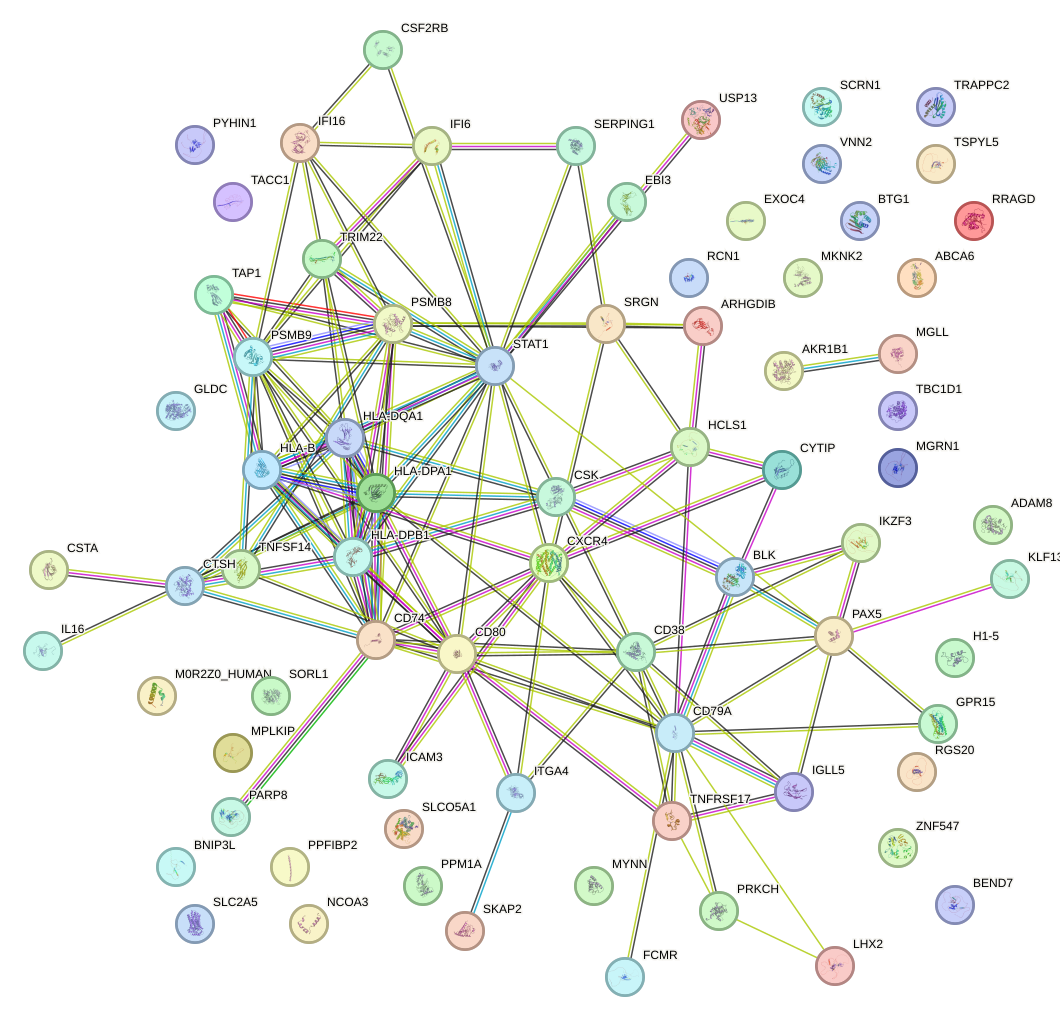
Network of associatons between targets according to the STRING database.
First level regulatory network of NR3C1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_106054659 | 7.27 |
ENST00000390539.2 |
IGHA2 |
immunoglobulin heavy constant alpha 2 (A2m marker) |
| chr14_-_106692191 | 7.07 |
ENST00000390607.2 |
IGHV3-21 |
immunoglobulin heavy variable 3-21 |
| chr22_+_23237555 | 5.85 |
ENST00000390321.2 |
IGLC1 |
immunoglobulin lambda constant 1 (Mcg marker) |
| chr22_+_23248512 | 5.49 |
ENST00000390325.2 |
IGLC3 |
immunoglobulin lambda constant 3 (Kern-Oz+ marker) |
| chr22_+_23243156 | 4.88 |
ENST00000390323.2 |
IGLC2 |
immunoglobulin lambda constant 2 (Kern-Oz- marker) |
| chr16_+_32077386 | 4.79 |
ENST00000354689.6 |
IGHV3OR16-9 |
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr14_-_106174960 | 4.67 |
ENST00000390547.2 |
IGHA1 |
immunoglobulin heavy constant alpha 1 |
| chr22_+_23241661 | 3.67 |
ENST00000390322.2 |
IGLJ2 |
immunoglobulin lambda joining 2 |
| chr6_-_33048483 | 3.44 |
ENST00000419277.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr6_+_33048222 | 3.42 |
ENST00000428835.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr5_-_149792295 | 3.03 |
ENST00000518797.1 ENST00000524315.1 ENST00000009530.7 ENST00000377795.3 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
| chr14_-_106994333 | 2.73 |
ENST00000390624.2 |
IGHV3-48 |
immunoglobulin heavy variable 3-48 |
| chr15_-_22448819 | 2.59 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr22_+_23264766 | 2.58 |
ENST00000390331.2 |
IGLC7 |
immunoglobulin lambda constant 7 |
| chr14_-_106518922 | 2.47 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr14_-_106642049 | 2.40 |
ENST00000390605.2 |
IGHV1-18 |
immunoglobulin heavy variable 1-18 |
| chr1_+_158900568 | 2.38 |
ENST00000458222.1 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr3_-_119278446 | 2.29 |
ENST00000264246.3 |
CD80 |
CD80 molecule |
| chr4_+_15779901 | 2.18 |
ENST00000226279.3 |
CD38 |
CD38 molecule |
| chr19_+_4229495 | 2.10 |
ENST00000221847.5 |
EBI3 |
Epstein-Barr virus induced 3 |
| chr2_-_158300556 | 2.10 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr22_+_23040274 | 2.02 |
ENST00000390306.2 |
IGLV2-23 |
immunoglobulin lambda variable 2-23 |
| chr14_-_107219365 | 1.99 |
ENST00000424969.2 |
IGHV3-74 |
immunoglobulin heavy variable 3-74 |
| chr14_-_106330458 | 1.99 |
ENST00000461719.1 |
IGHJ4 |
immunoglobulin heavy joining 4 |
| chr15_+_31658349 | 1.91 |
ENST00000558844.1 |
KLF13 |
Kruppel-like factor 13 |
| chr6_+_32605195 | 1.88 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr1_-_207095324 | 1.87 |
ENST00000530505.1 ENST00000367091.3 ENST00000442471.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr15_-_20193370 | 1.83 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr1_+_158969752 | 1.79 |
ENST00000566111.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr8_+_54793454 | 1.72 |
ENST00000276500.4 |
RGS20 |
regulator of G-protein signaling 20 |
| chr14_-_106573756 | 1.70 |
ENST00000390601.2 |
IGHV3-11 |
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr8_+_54793425 | 1.69 |
ENST00000522225.1 |
RGS20 |
regulator of G-protein signaling 20 |
| chr6_+_29795595 | 1.63 |
ENST00000360323.6 ENST00000376818.3 ENST00000376815.3 |
HLA-G |
major histocompatibility complex, class I, G |
| chr5_+_49961727 | 1.62 |
ENST00000505697.2 ENST00000503750.2 ENST00000514342.2 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_32811885 | 1.59 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr3_-_127541679 | 1.56 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr14_-_106668095 | 1.56 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr2_-_191878162 | 1.55 |
ENST00000540176.1 |
STAT1 |
signal transducer and activator of transcription 1, 91kDa |
| chr6_+_6588902 | 1.52 |
ENST00000230568.4 |
LY86 |
lymphocyte antigen 86 |
| chr3_+_122044084 | 1.51 |
ENST00000264474.3 ENST00000479204.1 |
CSTA |
cystatin A (stefin A) |
| chr14_-_106552755 | 1.50 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr14_-_106845789 | 1.45 |
ENST00000390617.2 |
IGHV3-35 |
immunoglobulin heavy variable 3-35 (non-functional) |
| chr11_+_121447469 | 1.45 |
ENST00000532694.1 ENST00000534286.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr6_+_32605134 | 1.45 |
ENST00000343139.5 ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr6_-_90121938 | 1.43 |
ENST00000369415.4 |
RRAGD |
Ras-related GTP binding D |
| chr14_-_106471723 | 1.42 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr14_-_106866934 | 1.41 |
ENST00000390618.2 |
IGHV3-38 |
immunoglobulin heavy variable 3-38 (non-functional) |
| chr6_-_31324943 | 1.40 |
ENST00000412585.2 ENST00000434333.1 |
HLA-B |
major histocompatibility complex, class I, B |
| chr1_-_207095212 | 1.39 |
ENST00000420007.2 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
| chr14_-_106453155 | 1.38 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr14_-_106926724 | 1.38 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr16_+_12058961 | 1.36 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr16_-_33647696 | 1.34 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr9_-_6645628 | 1.33 |
ENST00000321612.6 |
GLDC |
glycine dehydrogenase (decarboxylating) |
| chr3_-_127541194 | 1.32 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr8_+_11351876 | 1.32 |
ENST00000529894.1 |
BLK |
B lymphoid tyrosine kinase |
| chr6_-_32811771 | 1.29 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr14_-_107049312 | 1.26 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chr16_+_33020496 | 1.26 |
ENST00000565407.2 |
IGHV3OR16-8 |
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr14_-_106725723 | 1.26 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr11_+_5710919 | 1.25 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr19_+_42381173 | 1.24 |
ENST00000221972.3 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr8_-_70745575 | 1.22 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr6_+_32821924 | 1.21 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr12_-_15114492 | 1.21 |
ENST00000541546.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr11_+_7626950 | 1.21 |
ENST00000530181.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr20_+_46130619 | 1.20 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr15_-_79237433 | 1.13 |
ENST00000220166.5 |
CTSH |
cathepsin H |
| chr3_+_98250743 | 1.13 |
ENST00000284311.3 |
GPR15 |
G protein-coupled receptor 15 |
| chr1_-_9129598 | 1.13 |
ENST00000535586.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr6_-_133079022 | 1.12 |
ENST00000525289.1 ENST00000326499.6 |
VNN2 |
vanin 2 |
| chr22_+_22764088 | 1.11 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr7_-_30029574 | 1.10 |
ENST00000426154.1 ENST00000421434.1 ENST00000434476.2 |
SCRN1 |
secernin 1 |
| chr7_-_94285511 | 1.08 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr22_+_22712087 | 1.07 |
ENST00000390294.2 |
IGLV1-47 |
immunoglobulin lambda variable 1-47 |
| chr19_-_2041159 | 1.07 |
ENST00000589441.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr3_-_119278376 | 1.07 |
ENST00000478182.1 |
CD80 |
CD80 molecule |
| chr19_+_57874835 | 1.02 |
ENST00000543226.1 ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1 ZNF547 AC003002.4 |
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr7_-_134143841 | 1.02 |
ENST00000285930.4 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr1_-_27998689 | 1.02 |
ENST00000339145.4 ENST00000362020.4 ENST00000361157.6 |
IFI6 |
interferon, alpha-inducible protein 6 |
| chr5_+_67588391 | 1.01 |
ENST00000523872.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr7_-_94285402 | 0.97 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr9_-_37034028 | 0.97 |
ENST00000520281.1 ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5 |
paired box 5 |
| chr4_+_37892682 | 0.94 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr3_+_52245458 | 0.93 |
ENST00000459884.1 |
ALAS1 |
aminolevulinate, delta-, synthase 1 |
| chr8_-_98290087 | 0.93 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr9_-_88356789 | 0.93 |
ENST00000357081.3 ENST00000376081.4 ENST00000337006.4 ENST00000376109.3 |
AGTPBP1 |
ATP/GTP binding protein 1 |
| chr2_-_175499294 | 0.92 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr14_-_106733624 | 0.91 |
ENST00000390610.2 |
IGHV1-24 |
immunoglobulin heavy variable 1-24 |
| chr2_-_136875712 | 0.91 |
ENST00000241393.3 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
| chr7_-_30029367 | 0.91 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chr19_-_10445399 | 0.88 |
ENST00000592945.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr2_+_182321925 | 0.87 |
ENST00000339307.4 ENST00000397033.2 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chr22_-_17680472 | 0.87 |
ENST00000330232.4 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
| chr12_-_92539614 | 0.86 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr8_+_38614807 | 0.85 |
ENST00000330691.6 ENST00000348567.4 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr16_+_4674787 | 0.85 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr14_+_60715928 | 0.85 |
ENST00000395076.4 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr9_+_126777676 | 0.84 |
ENST00000488674.2 |
LHX2 |
LIM homeobox 2 |
| chr17_-_38020392 | 0.84 |
ENST00000346872.3 ENST00000439167.2 ENST00000377945.3 ENST00000394189.2 ENST00000377944.3 ENST00000377958.2 ENST00000535189.1 ENST00000377952.2 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
| chr15_+_81475047 | 0.84 |
ENST00000559388.1 |
IL16 |
interleukin 16 |
| chr15_+_25200108 | 0.83 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr21_-_46340884 | 0.83 |
ENST00000302347.5 ENST00000517819.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr21_-_46340770 | 0.82 |
ENST00000397854.3 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr3_+_179370517 | 0.81 |
ENST00000263966.3 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr6_-_109777128 | 0.77 |
ENST00000358807.3 ENST00000358577.3 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr6_-_32821599 | 0.76 |
ENST00000354258.4 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr22_+_22681656 | 0.75 |
ENST00000390291.2 |
IGLV1-50 |
immunoglobulin lambda variable 1-50 (non-functional) |
| chr8_+_26247878 | 0.75 |
ENST00000518611.1 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr14_+_61654271 | 0.73 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr19_-_39108552 | 0.73 |
ENST00000591517.1 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr2_-_89292422 | 0.72 |
ENST00000495489.1 |
IGKV1-8 |
immunoglobulin kappa variable 1-8 |
| chr4_-_105416039 | 0.72 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr14_-_107114267 | 0.71 |
ENST00000454421.2 |
IGHV3-64 |
immunoglobulin heavy variable 3-64 |
| chr3_-_121379739 | 0.70 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr7_+_26332645 | 0.70 |
ENST00000396376.1 |
SNX10 |
sorting nexin 10 |
| chr14_+_24641062 | 0.69 |
ENST00000311457.3 ENST00000557806.1 ENST00000559919.1 |
REC8 |
REC8 meiotic recombination protein |
| chr14_-_106816253 | 0.67 |
ENST00000390615.2 |
IGHV3-33 |
immunoglobulin heavy variable 3-33 |
| chr8_-_103136481 | 0.67 |
ENST00000524209.1 ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD |
neurocalcin delta |
| chr6_-_27835357 | 0.61 |
ENST00000331442.3 |
HIST1H1B |
histone cluster 1, H1b |
| chr22_+_37309662 | 0.60 |
ENST00000403662.3 ENST00000262825.5 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr7_-_40174201 | 0.60 |
ENST00000306984.6 |
MPLKIP |
M-phase specific PLK1 interacting protein |
| chr2_-_214016314 | 0.60 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr10_-_13523073 | 0.60 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr2_+_233925064 | 0.60 |
ENST00000359570.5 ENST00000538935.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr3_-_58200398 | 0.60 |
ENST00000318316.3 ENST00000460422.1 ENST00000483681.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr17_-_67138015 | 0.60 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr10_-_135090338 | 0.59 |
ENST00000415217.3 |
ADAM8 |
ADAM metallopeptidase domain 8 |
| chr16_+_4674814 | 0.59 |
ENST00000415496.1 ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr10_+_70847852 | 0.59 |
ENST00000242465.3 |
SRGN |
serglycin |
| chr11_+_57365150 | 0.59 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr19_-_6670128 | 0.59 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr15_+_75080883 | 0.58 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr2_-_89340242 | 0.58 |
ENST00000480492.1 |
IGKV1-12 |
immunoglobulin kappa variable 1-12 |
| chr16_+_33006369 | 0.57 |
ENST00000425181.3 |
IGHV3OR16-10 |
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr3_+_169490606 | 0.56 |
ENST00000349841.5 |
MYNN |
myoneurin |
| chr11_+_32112431 | 0.55 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr7_-_26904317 | 0.54 |
ENST00000345317.2 |
SKAP2 |
src kinase associated phosphoprotein 2 |
| chr11_+_117857063 | 0.53 |
ENST00000227752.3 ENST00000541785.1 ENST00000545409.1 |
IL10RA |
interleukin 10 receptor, alpha |
| chr18_+_34124507 | 0.52 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr2_-_89513402 | 0.52 |
ENST00000498435.1 |
IGKV1-27 |
immunoglobulin kappa variable 1-27 |
| chr22_-_37172111 | 0.52 |
ENST00000417951.2 ENST00000430701.1 ENST00000433985.2 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr2_+_102608306 | 0.50 |
ENST00000332549.3 |
IL1R2 |
interleukin 1 receptor, type II |
| chr20_-_62711259 | 0.50 |
ENST00000332298.5 |
RGS19 |
regulator of G-protein signaling 19 |
| chr16_+_50730910 | 0.50 |
ENST00000300589.2 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
| chr6_-_137113604 | 0.50 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr22_-_37172191 | 0.50 |
ENST00000340630.5 |
IFT27 |
intraflagellar transport 27 homolog (Chlamydomonas) |
| chr9_-_21335240 | 0.50 |
ENST00000537938.1 |
KLHL9 |
kelch-like family member 9 |
| chr16_+_84209539 | 0.50 |
ENST00000569735.1 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
| chr6_-_39693111 | 0.49 |
ENST00000373215.3 ENST00000538893.1 ENST00000287152.7 ENST00000373216.3 |
KIF6 |
kinesin family member 6 |
| chr17_-_34756219 | 0.49 |
ENST00000451448.2 ENST00000394359.3 |
TBC1D3C TBC1D3H |
TBC1 domain family, member 3C TBC1 domain family, member 3H |
| chr3_-_69101413 | 0.49 |
ENST00000398559.2 |
TMF1 |
TATA element modulatory factor 1 |
| chr16_+_32063311 | 0.49 |
ENST00000426099.1 |
AC142381.1 |
AC142381.1 |
| chr10_-_135090360 | 0.48 |
ENST00000486609.1 ENST00000445355.3 ENST00000485491.2 |
ADAM8 |
ADAM metallopeptidase domain 8 |
| chr18_+_13611763 | 0.48 |
ENST00000585931.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr15_+_74833518 | 0.48 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chr19_+_58281014 | 0.48 |
ENST00000391702.3 ENST00000598885.1 ENST00000598183.1 ENST00000396154.2 ENST00000599802.1 ENST00000396150.4 |
ZNF586 |
zinc finger protein 586 |
| chr3_-_69101461 | 0.47 |
ENST00000543976.1 |
TMF1 |
TATA element modulatory factor 1 |
| chr17_+_10600894 | 0.46 |
ENST00000379774.4 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr6_-_132834184 | 0.46 |
ENST00000367941.2 ENST00000367937.4 |
STX7 |
syntaxin 7 |
| chr14_-_23426270 | 0.46 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr14_+_77787227 | 0.45 |
ENST00000216465.5 ENST00000361389.4 ENST00000554279.1 ENST00000557639.1 ENST00000349555.3 ENST00000556627.1 ENST00000557053.1 |
GSTZ1 |
glutathione S-transferase zeta 1 |
| chr4_-_8442438 | 0.44 |
ENST00000356406.5 ENST00000413009.2 |
ACOX3 |
acyl-CoA oxidase 3, pristanoyl |
| chr12_+_66582919 | 0.44 |
ENST00000545837.1 ENST00000457197.2 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
| chr19_-_19774473 | 0.43 |
ENST00000357324.6 |
ATP13A1 |
ATPase type 13A1 |
| chr15_-_81616446 | 0.43 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr17_-_3599327 | 0.43 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr14_-_23426337 | 0.41 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chrX_+_2609207 | 0.41 |
ENST00000381192.3 |
CD99 |
CD99 molecule |
| chr6_-_26250835 | 0.41 |
ENST00000446824.2 |
HIST1H3F |
histone cluster 1, H3f |
| chr9_-_19127474 | 0.41 |
ENST00000380465.3 ENST00000380464.3 ENST00000411567.1 ENST00000276914.2 |
PLIN2 |
perilipin 2 |
| chr1_+_168195229 | 0.40 |
ENST00000271375.4 ENST00000367825.3 |
SFT2D2 |
SFT2 domain containing 2 |
| chr14_+_72064945 | 0.40 |
ENST00000537413.1 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr14_-_23426322 | 0.40 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr7_-_95225768 | 0.39 |
ENST00000005178.5 |
PDK4 |
pyruvate dehydrogenase kinase, isozyme 4 |
| chr7_-_22234381 | 0.39 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr6_-_33282163 | 0.39 |
ENST00000434618.2 ENST00000456592.2 |
TAPBP |
TAP binding protein (tapasin) |
| chr12_-_108714412 | 0.39 |
ENST00000412676.1 ENST00000550573.1 |
CMKLR1 |
chemokine-like receptor 1 |
| chr3_-_53880401 | 0.39 |
ENST00000315251.6 |
CHDH |
choline dehydrogenase |
| chr2_-_89521942 | 0.38 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr2_+_219110149 | 0.38 |
ENST00000456575.1 |
ARPC2 |
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr3_-_11623804 | 0.38 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr2_+_89998789 | 0.37 |
ENST00000453166.2 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr22_+_39410088 | 0.37 |
ENST00000361441.4 |
APOBEC3C |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3C |
| chr15_-_64126084 | 0.37 |
ENST00000560316.1 ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr12_-_498620 | 0.37 |
ENST00000399788.2 ENST00000382815.4 |
KDM5A |
lysine (K)-specific demethylase 5A |
| chr17_+_4843303 | 0.37 |
ENST00000571816.1 |
RNF167 |
ring finger protein 167 |
| chr11_-_85376121 | 0.37 |
ENST00000527447.1 |
CREBZF |
CREB/ATF bZIP transcription factor |
| chrX_+_2609317 | 0.37 |
ENST00000381187.3 ENST00000381184.1 |
CD99 |
CD99 molecule |
| chr20_+_43160409 | 0.37 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr10_+_120863587 | 0.36 |
ENST00000535029.1 ENST00000361432.2 ENST00000544016.1 |
FAM45A |
family with sequence similarity 45, member A |
| chr19_+_36157715 | 0.36 |
ENST00000379013.2 ENST00000222275.2 |
UPK1A |
uroplakin 1A |
| chr20_+_45947246 | 0.36 |
ENST00000599904.1 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr6_+_26045603 | 0.36 |
ENST00000540144.1 |
HIST1H3C |
histone cluster 1, H3c |
| chr3_-_66551397 | 0.36 |
ENST00000383703.3 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr19_+_8117881 | 0.35 |
ENST00000390669.3 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr3_-_119396193 | 0.35 |
ENST00000484810.1 ENST00000497116.1 ENST00000261070.2 |
COX17 |
COX17 cytochrome c oxidase copper chaperone |
| chr3_+_73110810 | 0.35 |
ENST00000533473.1 |
EBLN2 |
endogenous Bornavirus-like nucleoprotein 2 |
| chr5_+_125935960 | 0.35 |
ENST00000297540.4 |
PHAX |
phosphorylated adaptor for RNA export |
| chr2_-_70418032 | 0.35 |
ENST00000425268.1 ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42 |
chromosome 2 open reading frame 42 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 11.9 | GO:0071749 | IgA immunoglobulin complex(GO:0071745) IgA immunoglobulin complex, circulating(GO:0071746) monomeric IgA immunoglobulin complex(GO:0071748) polymeric IgA immunoglobulin complex(GO:0071749) secretory IgA immunoglobulin complex(GO:0071751) |
| 0.9 | 41.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.6 | 13.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 4.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.4 | 1.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.4 | 1.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.3 | 1.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.3 | 3.0 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.3 | 2.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.0 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.3 | 1.0 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.2 | 1.4 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.2 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.2 | 0.9 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.2 | 1.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.8 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.5 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 3.7 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.1 | GO:0005713 | recombination nodule(GO:0005713) |
| 0.1 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.5 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.3 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.1 | 1.0 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 9.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.7 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.7 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.2 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.6 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.3 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.2 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 3.5 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.0 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.6 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 3.0 | GO:0005769 | early endosome(GO:0005769) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 3.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 1.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 1.7 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 2.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 1.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.0 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 2.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 5.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.7 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 52.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.7 | 2.2 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.6 | 3.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.5 | 6.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.3 | 0.9 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.3 | 1.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.3 | 0.8 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.2 | 7.3 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.2 | 1.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.2 | 2.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 0.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 28.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.2 | 1.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.2 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 1.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.4 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 1.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 3.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0051538 | 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.1 | 0.3 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 1.3 | GO:0016594 | glycine binding(GO:0016594) |
| 0.1 | 0.5 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 1.5 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.1 | 2.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.4 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.1 | 4.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.3 | GO:0016160 | amylase activity(GO:0016160) maltose alpha-glucosidase activity(GO:0032450) |
| 0.1 | 3.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.4 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.1 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.5 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.8 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.4 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.7 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.5 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.7 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 1.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0015307 | drug:proton antiporter activity(GO:0015307) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 1.1 | GO:0016903 | oxidoreductase activity, acting on the aldehyde or oxo group of donors(GO:0016903) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004797 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.6 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 10.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.3 | 3.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.2 | 0.9 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 3.7 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 2.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 1.6 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 3.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 4.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 4.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.8 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 3.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | Genes involved in Assembly of the pre-replicative complex |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 11.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.6 | 3.0 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.6 | 41.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.5 | 1.6 | GO:0035419 | activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.5 | 1.5 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.5 | 3.4 | GO:0045425 | regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.4 | 2.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.4 | 1.6 | GO:2000697 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of nephron tubule epithelial cell differentiation(GO:0072183) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) negative regulation of epithelial cell differentiation involved in kidney development(GO:2000697) |
| 0.4 | 1.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.3 | 1.0 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 0.3 | 0.9 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 3.0 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.3 | 1.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 1.3 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.2 | 0.7 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 1.4 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.2 | 0.7 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.2 | 0.9 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.2 | 3.4 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.2 | 0.8 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 0.6 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 0.6 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.2 | 1.7 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.2 | 1.0 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.2 | 1.9 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 1.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 0.8 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.2 | 1.2 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.2 | 0.5 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.2 | 1.1 | GO:0015755 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.2 | 16.2 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 0.8 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.1 | 0.4 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.1 | 0.4 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.1 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.9 | GO:0019064 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.5 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.1 | 0.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.6 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.3 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.7 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 1.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 11.2 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.1 | 1.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) |
| 0.1 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.1 | 0.4 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 1.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.3 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.1 | 0.3 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.1 | 2.3 | GO:0050855 | regulation of B cell receptor signaling pathway(GO:0050855) |
| 0.1 | 0.2 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 0.8 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.2 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.1 | 0.3 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.7 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 1.0 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.7 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.1 | 0.6 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.9 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.1 | 0.2 | GO:0038060 | nitric oxide-cGMP-mediated signaling pathway(GO:0038060) |
| 0.1 | 0.3 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) |
| 0.1 | 0.3 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 0.4 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.2 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.5 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.1 | 1.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.3 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.3 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.4 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.8 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.0 | 0.8 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 2.0 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 1.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.4 | GO:0021702 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.7 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0046440 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.0 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.5 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.3 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.4 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.0 | 0.8 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.4 | GO:0050942 | positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.3 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 0.4 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.2 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 0.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.0 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 3.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 1.4 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.9 | GO:0072678 | T cell migration(GO:0072678) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.9 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.5 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 1.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.0 | 0.5 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.3 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.4 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:2000382 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 1.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.3 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.0 | 0.9 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 1.9 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.0 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.3 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.0 | 0.2 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 0.3 | GO:0002717 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.0 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.3 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 4.3 | GO:0002250 | adaptive immune response(GO:0002250) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0007588 | excretion(GO:0007588) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.0 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.0 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.2 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.0 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.0 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.2 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |