Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
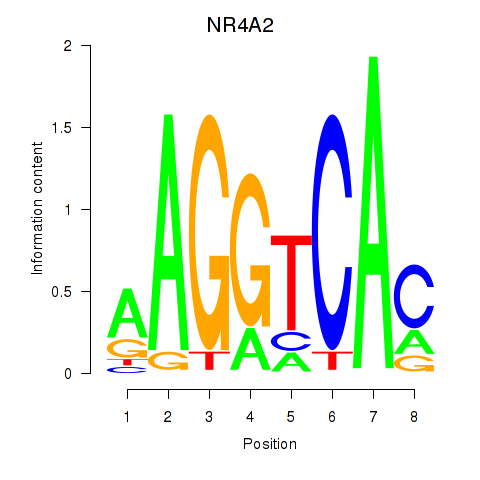
Results for NR4A2
Z-value: 0.86
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | NR4A2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A2 | hg19_v2_chr2_-_157198860_157198978 | 0.52 | 4.0e-02 | Click! |
Activity profile of NR4A2 motif
Sorted Z-values of NR4A2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43985725 | 1.83 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr15_+_43985084 | 1.79 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr1_-_17380630 | 1.71 |
ENST00000375499.3 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr15_+_43885252 | 1.66 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr10_+_81107271 | 1.42 |
ENST00000448165.1 |
PPIF |
peptidylprolyl isomerase F |
| chr8_-_131028869 | 1.39 |
ENST00000518283.1 ENST00000519110.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr3_-_167452262 | 1.31 |
ENST00000487947.2 |
PDCD10 |
programmed cell death 10 |
| chr15_-_55562479 | 1.27 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_+_93963590 | 1.23 |
ENST00000340600.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr9_-_138853156 | 1.12 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chrX_-_109683446 | 1.08 |
ENST00000372057.1 |
AMMECR1 |
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr19_+_45394477 | 1.07 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr2_+_234621551 | 1.04 |
ENST00000608381.1 ENST00000373414.3 |
UGT1A1 UGT1A5 |
UDP glucuronosyltransferase 1 family, polypeptide A8 UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr10_+_14880157 | 1.01 |
ENST00000378372.3 |
HSPA14 |
heat shock 70kDa protein 14 |
| chr11_+_66624527 | 1.00 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr3_-_151034734 | 0.96 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr1_-_241683001 | 0.95 |
ENST00000366560.3 |
FH |
fumarate hydratase |
| chr19_-_8008533 | 0.89 |
ENST00000597926.1 |
TIMM44 |
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr3_-_167452298 | 0.89 |
ENST00000475915.2 ENST00000462725.2 ENST00000461494.1 |
PDCD10 |
programmed cell death 10 |
| chr8_-_131028641 | 0.84 |
ENST00000523509.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr15_-_55562582 | 0.84 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr14_-_21492251 | 0.79 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_21492113 | 0.77 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr11_-_64013663 | 0.75 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr3_-_113465065 | 0.71 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_+_14989186 | 0.70 |
ENST00000435454.1 ENST00000323373.6 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr10_+_51371390 | 0.70 |
ENST00000478381.1 ENST00000451577.2 ENST00000374098.2 ENST00000374097.2 |
TIMM23B |
translocase of inner mitochondrial membrane 23 homolog B (yeast) |
| chr10_+_81107216 | 0.69 |
ENST00000394579.3 ENST00000225174.3 |
PPIF |
peptidylprolyl isomerase F |
| chr10_-_51623203 | 0.68 |
ENST00000444743.1 ENST00000374065.3 ENST00000374064.3 ENST00000260867.4 |
TIMM23 |
translocase of inner mitochondrial membrane 23 homolog (yeast) |
| chr11_-_64014379 | 0.67 |
ENST00000309318.3 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr15_+_75335604 | 0.66 |
ENST00000563393.1 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
| chr2_-_176046391 | 0.66 |
ENST00000392541.3 ENST00000409194.1 |
ATP5G3 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr1_+_201924619 | 0.65 |
ENST00000367287.4 |
TIMM17A |
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr10_+_22605374 | 0.65 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr2_+_85804614 | 0.63 |
ENST00000263864.5 ENST00000409760.1 |
VAMP8 |
vesicle-associated membrane protein 8 |
| chr19_+_8509842 | 0.62 |
ENST00000325495.4 ENST00000600092.1 ENST00000594907.1 ENST00000596984.1 ENST00000601645.1 |
HNRNPM |
heterogeneous nuclear ribonucleoprotein M |
| chr16_-_69368774 | 0.61 |
ENST00000562949.1 |
RP11-343C2.12 |
Conserved oligomeric Golgi complex subunit 8 |
| chr15_+_78441663 | 0.61 |
ENST00000299518.2 ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A |
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr14_-_104387888 | 0.59 |
ENST00000286953.3 |
C14orf2 |
chromosome 14 open reading frame 2 |
| chr10_+_22605304 | 0.58 |
ENST00000475460.2 ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1 COMMD3 |
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr19_-_2328572 | 0.57 |
ENST00000252622.10 |
LSM7 |
LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr19_-_46476791 | 0.55 |
ENST00000263257.5 |
NOVA2 |
neuro-oncological ventral antigen 2 |
| chr16_-_4292071 | 0.54 |
ENST00000399609.3 |
SRL |
sarcalumenin |
| chr14_-_104387831 | 0.54 |
ENST00000557040.1 ENST00000414262.2 ENST00000555030.1 ENST00000554713.1 ENST00000553430.1 |
C14orf2 |
chromosome 14 open reading frame 2 |
| chr7_-_140624499 | 0.53 |
ENST00000288602.6 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr8_+_145149930 | 0.53 |
ENST00000318911.4 |
CYC1 |
cytochrome c-1 |
| chr1_+_196621156 | 0.51 |
ENST00000359637.2 |
CFH |
complement factor H |
| chr1_+_206858328 | 0.49 |
ENST00000367103.3 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
| chr12_+_120875910 | 0.49 |
ENST00000551806.1 |
AL021546.6 |
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr19_-_10446449 | 0.48 |
ENST00000592439.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr22_-_27620603 | 0.48 |
ENST00000418271.1 ENST00000444114.1 |
RP5-1172A22.1 |
RP5-1172A22.1 |
| chr22_+_31518938 | 0.47 |
ENST00000412985.1 ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr20_-_60718430 | 0.47 |
ENST00000370873.4 ENST00000370858.3 |
PSMA7 |
proteasome (prosome, macropain) subunit, alpha type, 7 |
| chr2_+_234627424 | 0.47 |
ENST00000373409.3 |
UGT1A4 |
UDP glucuronosyltransferase 1 family, polypeptide A4 |
| chr11_-_66139199 | 0.47 |
ENST00000357440.2 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
| chr8_-_70745575 | 0.46 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr17_+_79670386 | 0.45 |
ENST00000333676.3 ENST00000571730.1 ENST00000541223.1 |
MRPL12 SLC25A10 SLC25A10 |
mitochondrial ribosomal protein L12 Mitochondrial dicarboxylate carrier; Uncharacterized protein; cDNA FLJ60124, highly similar to Mitochondrial dicarboxylate carrier solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr7_-_91875109 | 0.43 |
ENST00000412043.2 ENST00000430102.1 ENST00000425073.1 ENST00000394503.2 ENST00000454017.1 ENST00000440209.1 ENST00000413688.1 ENST00000452773.1 ENST00000433016.1 ENST00000394505.2 ENST00000422347.1 ENST00000458493.1 ENST00000425919.1 |
KRIT1 |
KRIT1, ankyrin repeat containing |
| chr11_-_64511575 | 0.42 |
ENST00000431822.1 ENST00000377486.3 ENST00000394432.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr1_-_1711508 | 0.42 |
ENST00000378625.1 |
NADK |
NAD kinase |
| chr4_+_20255123 | 0.41 |
ENST00000504154.1 ENST00000273739.5 |
SLIT2 |
slit homolog 2 (Drosophila) |
| chr12_-_120554534 | 0.41 |
ENST00000538903.1 ENST00000534951.1 |
RAB35 |
RAB35, member RAS oncogene family |
| chr6_+_107077435 | 0.41 |
ENST00000369046.4 |
QRSL1 |
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr20_-_1373606 | 0.40 |
ENST00000381715.1 ENST00000439640.2 ENST00000381719.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr10_+_11207438 | 0.40 |
ENST00000609692.1 ENST00000354897.3 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chrX_-_63005405 | 0.40 |
ENST00000374878.1 ENST00000437457.2 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr1_+_32757668 | 0.39 |
ENST00000373548.3 |
HDAC1 |
histone deacetylase 1 |
| chr22_+_30792980 | 0.39 |
ENST00000403484.1 ENST00000405717.3 ENST00000402592.3 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr4_-_71705060 | 0.39 |
ENST00000514161.1 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr1_+_196621002 | 0.39 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr4_-_71705027 | 0.39 |
ENST00000545193.1 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr20_-_62130474 | 0.39 |
ENST00000217182.3 |
EEF1A2 |
eukaryotic translation elongation factor 1 alpha 2 |
| chr20_+_57875658 | 0.38 |
ENST00000371025.3 |
EDN3 |
endothelin 3 |
| chr1_+_26348259 | 0.38 |
ENST00000374280.3 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
| chr4_-_71705082 | 0.37 |
ENST00000439371.1 ENST00000499044.2 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr4_-_151936416 | 0.36 |
ENST00000510413.1 ENST00000507224.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr3_+_113465866 | 0.36 |
ENST00000273398.3 ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr11_-_64512273 | 0.36 |
ENST00000377497.3 ENST00000377487.1 ENST00000430645.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr4_-_40517984 | 0.35 |
ENST00000381795.6 |
RBM47 |
RNA binding motif protein 47 |
| chr6_-_34664612 | 0.34 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr6_-_31628512 | 0.34 |
ENST00000375911.1 |
C6orf47 |
chromosome 6 open reading frame 47 |
| chr5_-_133340326 | 0.33 |
ENST00000425992.1 ENST00000395044.3 ENST00000395047.2 |
VDAC1 |
voltage-dependent anion channel 1 |
| chr3_-_197024965 | 0.33 |
ENST00000392382.2 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr1_+_16062820 | 0.33 |
ENST00000294454.5 |
SLC25A34 |
solute carrier family 25, member 34 |
| chr12_+_58013693 | 0.33 |
ENST00000320442.4 ENST00000379218.2 |
SLC26A10 |
solute carrier family 26, member 10 |
| chr1_+_206858232 | 0.33 |
ENST00000294981.4 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
| chr20_+_57875457 | 0.32 |
ENST00000337938.2 ENST00000311585.7 ENST00000371028.2 |
EDN3 |
endothelin 3 |
| chr7_-_91875358 | 0.32 |
ENST00000458177.1 ENST00000394507.1 ENST00000340022.2 ENST00000444960.1 |
KRIT1 |
KRIT1, ankyrin repeat containing |
| chr3_-_10028366 | 0.32 |
ENST00000429759.1 |
EMC3 |
ER membrane protein complex subunit 3 |
| chr1_+_155583012 | 0.31 |
ENST00000462250.2 |
MSTO1 |
misato 1, mitochondrial distribution and morphology regulator |
| chr11_+_67798363 | 0.31 |
ENST00000525628.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr4_-_151936865 | 0.31 |
ENST00000535741.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr19_+_589893 | 0.31 |
ENST00000251287.2 |
HCN2 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr20_+_57875758 | 0.30 |
ENST00000395654.3 |
EDN3 |
endothelin 3 |
| chr2_+_11817713 | 0.29 |
ENST00000449576.2 |
LPIN1 |
lipin 1 |
| chr8_-_100905925 | 0.29 |
ENST00000518171.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr3_+_159570722 | 0.28 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr11_-_19223523 | 0.28 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chrX_+_44732757 | 0.26 |
ENST00000377967.4 ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A |
lysine (K)-specific demethylase 6A |
| chr6_+_149068464 | 0.26 |
ENST00000367463.4 |
UST |
uronyl-2-sulfotransferase |
| chr1_-_29508499 | 0.25 |
ENST00000373795.4 |
SRSF4 |
serine/arginine-rich splicing factor 4 |
| chr7_-_72936531 | 0.24 |
ENST00000339594.4 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
| chr8_-_145018905 | 0.24 |
ENST00000398774.2 |
PLEC |
plectin |
| chr7_-_151433393 | 0.24 |
ENST00000492843.1 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr17_+_79679299 | 0.24 |
ENST00000331531.5 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr16_-_4466565 | 0.24 |
ENST00000572467.1 ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16 CORO7 |
CORO7-PAM16 readthrough coronin 7 |
| chr3_+_179322573 | 0.23 |
ENST00000493866.1 ENST00000472629.1 ENST00000482604.1 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr4_+_81118647 | 0.22 |
ENST00000415738.2 |
PRDM8 |
PR domain containing 8 |
| chr1_+_68150744 | 0.22 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr8_-_100905850 | 0.22 |
ENST00000520271.1 ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr3_+_159557637 | 0.22 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr7_-_151433342 | 0.21 |
ENST00000433631.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr6_+_136172820 | 0.21 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr11_-_84634217 | 0.21 |
ENST00000524982.1 |
DLG2 |
discs, large homolog 2 (Drosophila) |
| chr19_+_46850251 | 0.21 |
ENST00000012443.4 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr3_+_138340067 | 0.21 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr15_-_80215984 | 0.21 |
ENST00000485386.1 ENST00000479961.1 |
ST20 ST20-MTHFS |
suppressor of tumorigenicity 20 ST20-MTHFS readthrough |
| chr22_+_40573921 | 0.21 |
ENST00000454349.2 ENST00000335727.9 |
TNRC6B |
trinucleotide repeat containing 6B |
| chr17_+_79679369 | 0.21 |
ENST00000350690.5 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr22_+_18121562 | 0.21 |
ENST00000355028.3 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
| chr16_-_85833109 | 0.21 |
ENST00000253457.3 |
EMC8 |
ER membrane protein complex subunit 8 |
| chr16_-_30537839 | 0.20 |
ENST00000380412.5 |
ZNF768 |
zinc finger protein 768 |
| chr1_-_20141763 | 0.20 |
ENST00000375121.2 |
RNF186 |
ring finger protein 186 |
| chr1_+_155179012 | 0.20 |
ENST00000609421.1 |
MTX1 |
metaxin 1 |
| chr3_+_138340049 | 0.20 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr3_-_52860850 | 0.20 |
ENST00000441637.2 |
ITIH4 |
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr22_-_37880543 | 0.19 |
ENST00000442496.1 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr11_+_3876859 | 0.19 |
ENST00000300737.4 |
STIM1 |
stromal interaction molecule 1 |
| chr8_-_30706608 | 0.19 |
ENST00000256246.2 |
TEX15 |
testis expressed 15 |
| chr22_-_29784519 | 0.19 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr6_+_139456226 | 0.18 |
ENST00000367658.2 |
HECA |
headcase homolog (Drosophila) |
| chr22_+_30163340 | 0.18 |
ENST00000330029.6 ENST00000401406.3 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr14_+_22320634 | 0.17 |
ENST00000390435.1 |
TRAV8-3 |
T cell receptor alpha variable 8-3 |
| chr8_+_134203303 | 0.17 |
ENST00000519433.1 ENST00000517423.1 ENST00000377863.2 ENST00000220856.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
| chr7_+_45613958 | 0.16 |
ENST00000297323.7 |
ADCY1 |
adenylate cyclase 1 (brain) |
| chr14_+_22963806 | 0.16 |
ENST00000390493.1 |
TRAJ44 |
T cell receptor alpha joining 44 |
| chr2_+_191334212 | 0.16 |
ENST00000444317.1 ENST00000535751.1 |
MFSD6 |
major facilitator superfamily domain containing 6 |
| chr20_+_43343886 | 0.16 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr1_+_43803475 | 0.16 |
ENST00000372470.3 ENST00000413998.2 |
MPL |
myeloproliferative leukemia virus oncogene |
| chr9_+_113431059 | 0.16 |
ENST00000416899.2 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
| chr3_+_179322481 | 0.15 |
ENST00000259037.3 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr1_-_154164534 | 0.15 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr22_-_43036607 | 0.15 |
ENST00000505920.1 |
ATP5L2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr1_-_226129189 | 0.15 |
ENST00000366820.5 |
LEFTY2 |
left-right determination factor 2 |
| chr3_+_51741072 | 0.15 |
ENST00000395052.3 |
GRM2 |
glutamate receptor, metabotropic 2 |
| chr12_-_120554622 | 0.15 |
ENST00000229340.5 |
RAB35 |
RAB35, member RAS oncogene family |
| chr12_-_108954933 | 0.15 |
ENST00000431469.2 ENST00000546815.1 |
SART3 |
squamous cell carcinoma antigen recognized by T cells 3 |
| chr19_-_46272106 | 0.15 |
ENST00000560168.1 |
SIX5 |
SIX homeobox 5 |
| chr5_+_218356 | 0.14 |
ENST00000264932.6 ENST00000504309.1 ENST00000510361.1 |
SDHA |
succinate dehydrogenase complex, subunit A, flavoprotein (Fp) |
| chr3_-_15540055 | 0.14 |
ENST00000605797.1 ENST00000435459.2 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr5_-_137878887 | 0.14 |
ENST00000507939.1 ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1 |
eukaryotic translation termination factor 1 |
| chr8_-_100905363 | 0.13 |
ENST00000524245.1 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr21_+_35014706 | 0.13 |
ENST00000399353.1 ENST00000444491.1 ENST00000381318.3 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
| chr2_+_119981384 | 0.13 |
ENST00000393108.2 ENST00000354888.5 ENST00000450943.2 ENST00000393110.2 ENST00000393106.2 ENST00000409811.1 ENST00000393107.2 |
STEAP3 |
STEAP family member 3, metalloreductase |
| chr9_-_130487143 | 0.13 |
ENST00000419060.1 |
PTRH1 |
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr8_+_145582633 | 0.13 |
ENST00000540505.1 |
SLC52A2 |
solute carrier family 52 (riboflavin transporter), member 2 |
| chr11_-_64511789 | 0.13 |
ENST00000419843.1 ENST00000394430.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr6_-_24666819 | 0.13 |
ENST00000341060.3 |
TDP2 |
tyrosyl-DNA phosphodiesterase 2 |
| chr11_+_122709200 | 0.12 |
ENST00000227348.4 |
CRTAM |
cytotoxic and regulatory T cell molecule |
| chr1_-_226129083 | 0.12 |
ENST00000420304.2 |
LEFTY2 |
left-right determination factor 2 |
| chr6_+_30594619 | 0.12 |
ENST00000318999.7 ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
| chr5_+_66124590 | 0.11 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_150777949 | 0.11 |
ENST00000482571.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr18_-_12377283 | 0.11 |
ENST00000269143.3 |
AFG3L2 |
AFG3-like AAA ATPase 2 |
| chr19_-_49552006 | 0.11 |
ENST00000391869.3 |
CGB1 |
chorionic gonadotropin, beta polypeptide 1 |
| chr14_+_77228532 | 0.11 |
ENST00000167106.4 ENST00000554237.1 |
VASH1 |
vasohibin 1 |
| chr10_-_33625154 | 0.11 |
ENST00000265371.4 |
NRP1 |
neuropilin 1 |
| chr12_-_16761007 | 0.11 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr7_-_150777874 | 0.10 |
ENST00000540185.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr9_+_113431029 | 0.10 |
ENST00000189978.5 ENST00000374448.4 ENST00000374440.3 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
| chr17_-_56358287 | 0.10 |
ENST00000225275.3 ENST00000340482.3 |
MPO |
myeloperoxidase |
| chr19_+_6739662 | 0.10 |
ENST00000313285.8 ENST00000313244.9 ENST00000596758.1 |
TRIP10 |
thyroid hormone receptor interactor 10 |
| chr6_-_24667180 | 0.10 |
ENST00000545995.1 |
TDP2 |
tyrosyl-DNA phosphodiesterase 2 |
| chr17_-_32484313 | 0.10 |
ENST00000359872.6 |
ASIC2 |
acid-sensing (proton-gated) ion channel 2 |
| chr17_-_29624343 | 0.10 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr6_-_24489842 | 0.10 |
ENST00000230036.1 |
GPLD1 |
glycosylphosphatidylinositol specific phospholipase D1 |
| chr12_-_57039739 | 0.10 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr12_-_108955070 | 0.09 |
ENST00000228284.3 ENST00000546611.1 |
SART3 |
squamous cell carcinoma antigen recognized by T cells 3 |
| chr17_-_2169425 | 0.09 |
ENST00000570606.1 ENST00000354901.4 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr21_-_35014027 | 0.09 |
ENST00000399442.1 ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chr16_-_66952742 | 0.09 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr1_+_32084641 | 0.09 |
ENST00000373706.5 |
HCRTR1 |
hypocretin (orexin) receptor 1 |
| chr7_-_150777920 | 0.08 |
ENST00000353841.2 ENST00000297532.6 |
FASTK |
Fas-activated serine/threonine kinase |
| chr12_+_6309963 | 0.08 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr3_+_129247479 | 0.08 |
ENST00000296271.3 |
RHO |
rhodopsin |
| chr20_-_44485835 | 0.08 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr16_+_85942594 | 0.08 |
ENST00000566369.1 |
IRF8 |
interferon regulatory factor 8 |
| chr5_+_149569520 | 0.08 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr7_-_140179276 | 0.07 |
ENST00000443720.2 ENST00000255977.2 |
MKRN1 |
makorin ring finger protein 1 |
| chr13_+_112721913 | 0.07 |
ENST00000330949.1 |
SOX1 |
SRY (sex determining region Y)-box 1 |
| chr6_-_87804815 | 0.07 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr17_+_45973516 | 0.07 |
ENST00000376741.4 |
SP2 |
Sp2 transcription factor |
| chr19_+_54606145 | 0.07 |
ENST00000485876.1 ENST00000391762.1 ENST00000471292.1 ENST00000391763.3 ENST00000391764.3 ENST00000303553.5 |
NDUFA3 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa |
| chr14_-_25103388 | 0.06 |
ENST00000526004.1 ENST00000415355.3 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
| chr1_-_27816641 | 0.06 |
ENST00000430629.2 |
WASF2 |
WAS protein family, member 2 |
| chr16_-_49698136 | 0.06 |
ENST00000535559.1 |
ZNF423 |
zinc finger protein 423 |
| chr11_+_67798114 | 0.06 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr11_-_790060 | 0.06 |
ENST00000330106.4 |
CEND1 |
cell cycle exit and neuronal differentiation 1 |
| chr9_-_34048873 | 0.06 |
ENST00000449054.1 ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2 |
ubiquitin associated protein 2 |
| chr15_-_41120896 | 0.05 |
ENST00000299174.5 ENST00000427255.2 |
PPP1R14D |
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 4.3 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 1.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 5.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.2 | 2.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 2.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.2 | 1.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.7 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 2.9 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 0.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 0.4 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.1 | 0.6 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.7 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 1.0 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.5 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.4 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0045239 | tricarboxylic acid cycle enzyme complex(GO:0045239) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.8 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.1 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 5.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.2 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.5 | 3.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.4 | 2.2 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.3 | 0.9 | GO:0015131 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.3 | 3.1 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.2 | 1.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.2 | 1.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.5 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.2 | 0.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 2.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.3 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.3 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 2.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.1 | 0.4 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.2 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.6 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.9 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.5 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.5 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.9 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.5 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) |
| 0.0 | 0.1 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.8 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.4 | 2.2 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.4 | 2.2 | GO:2000568 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.4 | 2.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.3 | 1.0 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.3 | 3.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.3 | 1.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.3 | 0.9 | GO:1902356 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.3 | 4.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.3 | 1.5 | GO:0006789 | bilirubin conjugation(GO:0006789) |
| 0.3 | 1.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.2 | 0.6 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.2 | 1.9 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 0.4 | GO:0021966 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) corticospinal neuron axon guidance(GO:0021966) regulation of negative chemotaxis(GO:0050923) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.4 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.5 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 1.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.7 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.3 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.1 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.1 | 0.4 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.2 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.3 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.1 | 0.7 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.3 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.6 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.4 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.7 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.5 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.0 | 1.0 | GO:0015988 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.4 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.8 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.0 | 0.5 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.2 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 1.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.6 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |