Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
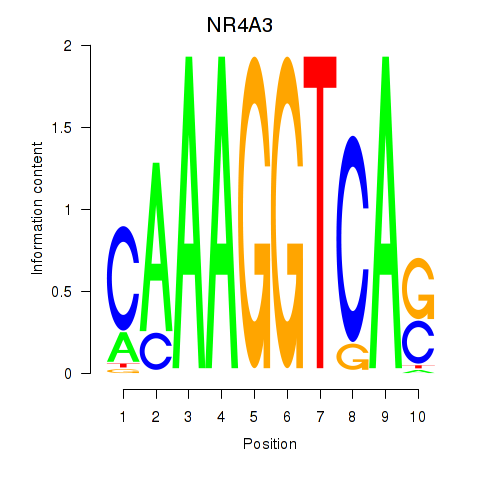
Results for NR4A3
Z-value: 0.96
Transcription factors associated with NR4A3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A3
|
ENSG00000119508.13 | NR4A3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR4A3 | hg19_v2_chr9_+_102584128_102584144 | -0.46 | 7.4e-02 | Click! |
Activity profile of NR4A3 motif
Sorted Z-values of NR4A3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR4A3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_228678550 | 4.07 |
ENST00000409189.3 ENST00000358813.4 |
CCL20 |
chemokine (C-C motif) ligand 20 |
| chr22_+_21128167 | 2.94 |
ENST00000215727.5 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr2_-_21266935 | 2.91 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chr17_-_64216748 | 2.37 |
ENST00000585162.1 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr13_+_113760098 | 2.23 |
ENST00000346342.3 ENST00000541084.1 ENST00000375581.3 |
F7 |
coagulation factor VII (serum prothrombin conversion accelerator) |
| chr1_-_169555779 | 1.94 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr12_-_122296755 | 1.86 |
ENST00000289004.4 |
HPD |
4-hydroxyphenylpyruvate dioxygenase |
| chr14_+_95047744 | 1.81 |
ENST00000553511.1 ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr9_-_75567962 | 1.78 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr1_+_64059332 | 1.73 |
ENST00000540265.1 |
PGM1 |
phosphoglucomutase 1 |
| chr17_+_4675175 | 1.71 |
ENST00000270560.3 |
TM4SF5 |
transmembrane 4 L six family member 5 |
| chr5_-_42811986 | 1.69 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr4_+_100495864 | 1.56 |
ENST00000265517.5 ENST00000422897.2 |
MTTP |
microsomal triglyceride transfer protein |
| chr13_-_46679144 | 1.55 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr13_-_46679185 | 1.53 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr10_-_125851961 | 1.49 |
ENST00000346248.5 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr15_+_58724184 | 1.43 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr11_+_22696314 | 1.40 |
ENST00000532398.1 ENST00000433790.1 |
GAS2 |
growth arrest-specific 2 |
| chrX_-_43741594 | 1.33 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr1_-_173886491 | 1.27 |
ENST00000367698.3 |
SERPINC1 |
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr16_+_72090053 | 1.26 |
ENST00000576168.2 ENST00000567185.3 ENST00000567612.2 |
HP |
haptoglobin |
| chr2_-_238499303 | 1.23 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr18_+_55816546 | 1.23 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr1_-_27240455 | 1.22 |
ENST00000254227.3 |
NR0B2 |
nuclear receptor subfamily 0, group B, member 2 |
| chr17_+_4854375 | 1.22 |
ENST00000521811.1 ENST00000519602.1 ENST00000323997.6 ENST00000522249.1 ENST00000519584.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr4_+_155484155 | 1.15 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr5_-_42812143 | 1.15 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr2_+_17721920 | 1.12 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr17_-_7081435 | 1.11 |
ENST00000380920.4 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr1_-_229569834 | 1.10 |
ENST00000366684.3 ENST00000366683.2 |
ACTA1 |
actin, alpha 1, skeletal muscle |
| chr18_+_55888767 | 1.09 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr10_+_101542462 | 1.08 |
ENST00000370449.4 ENST00000370434.1 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr3_-_50340996 | 1.08 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr2_+_128175997 | 1.04 |
ENST00000234071.3 ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr9_+_103790991 | 1.01 |
ENST00000374874.3 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chrX_+_38420623 | 1.01 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr20_+_61287711 | 0.99 |
ENST00000370507.1 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
| chr1_-_116311402 | 0.98 |
ENST00000261448.5 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
| chr14_+_32546274 | 0.98 |
ENST00000396582.2 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr5_-_95158644 | 0.96 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr4_-_16077741 | 0.95 |
ENST00000447510.2 ENST00000540805.1 ENST00000539194.1 |
PROM1 |
prominin 1 |
| chr7_-_87342564 | 0.95 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr7_-_87104963 | 0.94 |
ENST00000359206.3 ENST00000358400.3 ENST00000265723.4 |
ABCB4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr19_+_35773242 | 0.93 |
ENST00000222304.3 |
HAMP |
hepcidin antimicrobial peptide |
| chr8_+_11534462 | 0.92 |
ENST00000528712.1 ENST00000532977.1 |
GATA4 |
GATA binding protein 4 |
| chr4_-_70080449 | 0.91 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr7_+_7222233 | 0.91 |
ENST00000436587.2 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr17_+_4855053 | 0.90 |
ENST00000518175.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr11_-_57282349 | 0.89 |
ENST00000528450.1 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr2_+_170366203 | 0.88 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chrX_+_23801280 | 0.84 |
ENST00000379251.3 ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1 |
spermidine/spermine N1-acetyltransferase 1 |
| chr17_+_1646130 | 0.84 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr19_-_41256207 | 0.84 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr7_-_95025661 | 0.76 |
ENST00000542556.1 ENST00000265627.5 ENST00000427422.1 ENST00000451904.1 |
PON1 PON3 |
paraoxonase 1 paraoxonase 3 |
| chr5_-_107703556 | 0.75 |
ENST00000496714.1 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
| chr11_+_7618413 | 0.75 |
ENST00000528883.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr4_+_124317940 | 0.73 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr1_+_210502238 | 0.72 |
ENST00000545154.1 ENST00000537898.1 ENST00000391905.3 ENST00000545781.1 ENST00000261458.3 ENST00000308852.6 |
HHAT |
hedgehog acyltransferase |
| chr16_-_10652993 | 0.72 |
ENST00000536829.1 |
EMP2 |
epithelial membrane protein 2 |
| chr12_-_117537240 | 0.72 |
ENST00000392545.4 ENST00000541210.1 ENST00000335209.7 |
TESC |
tescalcin |
| chr13_-_52585547 | 0.71 |
ENST00000448424.2 ENST00000400370.3 ENST00000418097.2 ENST00000242839.4 ENST00000400366.3 ENST00000344297.5 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
| chr19_-_7293942 | 0.70 |
ENST00000341500.5 ENST00000302850.5 |
INSR |
insulin receptor |
| chr15_-_75017711 | 0.70 |
ENST00000567032.1 ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr2_-_238499725 | 0.70 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr19_+_35630628 | 0.69 |
ENST00000588715.1 ENST00000588607.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr12_-_8088871 | 0.69 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr16_-_87970122 | 0.68 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr11_-_70507901 | 0.67 |
ENST00000449833.2 ENST00000357171.3 ENST00000449116.2 |
SHANK2 |
SH3 and multiple ankyrin repeat domains 2 |
| chr18_+_29077990 | 0.66 |
ENST00000261590.8 |
DSG2 |
desmoglein 2 |
| chr10_-_116286563 | 0.66 |
ENST00000369253.2 |
ABLIM1 |
actin binding LIM protein 1 |
| chr9_-_95896550 | 0.66 |
ENST00000375446.4 |
NINJ1 |
ninjurin 1 |
| chr20_-_17641097 | 0.66 |
ENST00000246043.4 |
RRBP1 |
ribosome binding protein 1 |
| chrX_-_128657457 | 0.65 |
ENST00000371121.3 ENST00000371123.1 ENST00000371122.4 |
SMARCA1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr10_-_116444371 | 0.64 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr19_-_4535233 | 0.62 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr16_-_73082274 | 0.62 |
ENST00000268489.5 |
ZFHX3 |
zinc finger homeobox 3 |
| chr14_+_74035763 | 0.61 |
ENST00000238651.5 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr17_-_79995553 | 0.61 |
ENST00000581584.1 ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR |
dicarbonyl/L-xylulose reductase |
| chr19_-_15236470 | 0.60 |
ENST00000533747.1 ENST00000598709.1 ENST00000534378.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr9_-_94124171 | 0.60 |
ENST00000422391.2 ENST00000375731.4 ENST00000303617.5 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
| chr14_+_97059070 | 0.60 |
ENST00000553378.1 ENST00000555496.1 |
RP11-433J8.1 |
RP11-433J8.1 |
| chr4_-_111563076 | 0.60 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr17_-_10325261 | 0.60 |
ENST00000403437.2 |
MYH8 |
myosin, heavy chain 8, skeletal muscle, perinatal |
| chr2_-_238499131 | 0.59 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr15_+_66585555 | 0.58 |
ENST00000319194.5 ENST00000525134.2 ENST00000441424.2 |
DIS3L |
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr11_-_117699413 | 0.57 |
ENST00000528014.1 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr14_-_24610779 | 0.57 |
ENST00000560403.1 ENST00000419198.2 ENST00000216799.4 |
EMC9 |
ER membrane protein complex subunit 9 |
| chr19_-_15236562 | 0.57 |
ENST00000263383.3 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr17_-_34329084 | 0.55 |
ENST00000354059.4 ENST00000536149.1 |
CCL15 CCL14 |
chemokine (C-C motif) ligand 15 chemokine (C-C motif) ligand 14 |
| chr2_+_234296792 | 0.53 |
ENST00000409813.3 |
DGKD |
diacylglycerol kinase, delta 130kDa |
| chr11_-_77734260 | 0.53 |
ENST00000353172.5 |
KCTD14 |
potassium channel tetramerization domain containing 14 |
| chr3_+_128968437 | 0.52 |
ENST00000314797.6 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
| chr19_+_42301079 | 0.50 |
ENST00000596544.1 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr12_-_47219733 | 0.50 |
ENST00000547477.1 ENST00000447411.1 ENST00000266579.4 |
SLC38A4 |
solute carrier family 38, member 4 |
| chrX_+_46937745 | 0.49 |
ENST00000397180.1 ENST00000457380.1 ENST00000352078.4 |
RGN |
regucalcin |
| chr19_+_926000 | 0.48 |
ENST00000263620.3 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_-_117698787 | 0.47 |
ENST00000260287.2 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr11_-_117698765 | 0.47 |
ENST00000532119.1 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr12_+_109577202 | 0.47 |
ENST00000377848.3 ENST00000377854.5 |
ACACB |
acetyl-CoA carboxylase beta |
| chr22_-_30960876 | 0.46 |
ENST00000401975.1 ENST00000428682.1 ENST00000423299.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr10_+_80828774 | 0.46 |
ENST00000334512.5 |
ZMIZ1 |
zinc finger, MIZ-type containing 1 |
| chr20_+_35807512 | 0.44 |
ENST00000373622.5 |
RPN2 |
ribophorin II |
| chr2_-_238499337 | 0.43 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr1_+_244998602 | 0.43 |
ENST00000411948.2 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr3_-_178969403 | 0.43 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr11_+_17741111 | 0.42 |
ENST00000250003.3 |
MYOD1 |
myogenic differentiation 1 |
| chr22_-_39239987 | 0.42 |
ENST00000333039.2 |
NPTXR |
neuronal pentraxin receptor |
| chr17_+_55173933 | 0.42 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr1_-_20306909 | 0.41 |
ENST00000375111.3 ENST00000400520.3 |
PLA2G2A |
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr1_+_200993071 | 0.40 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr14_-_24911868 | 0.40 |
ENST00000554698.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr6_+_64346386 | 0.39 |
ENST00000509330.1 |
PHF3 |
PHD finger protein 3 |
| chr12_+_81101277 | 0.39 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr1_-_9811600 | 0.39 |
ENST00000435891.1 |
CLSTN1 |
calsyntenin 1 |
| chr22_+_45072925 | 0.39 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr22_+_45072958 | 0.38 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr7_-_143599207 | 0.38 |
ENST00000355951.2 ENST00000479870.1 ENST00000478172.1 |
FAM115A |
family with sequence similarity 115, member A |
| chr7_+_73868120 | 0.38 |
ENST00000265755.3 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr1_+_182992545 | 0.38 |
ENST00000258341.4 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
| chr17_+_77893135 | 0.37 |
ENST00000574526.1 ENST00000572353.1 |
RP11-353N14.4 |
RP11-353N14.4 |
| chr18_+_11981547 | 0.37 |
ENST00000588927.1 |
IMPA2 |
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr1_+_160097462 | 0.37 |
ENST00000447527.1 |
ATP1A2 |
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr2_+_121103706 | 0.36 |
ENST00000295228.3 |
INHBB |
inhibin, beta B |
| chr14_-_105262055 | 0.36 |
ENST00000349310.3 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr7_-_27169801 | 0.36 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr9_+_115983808 | 0.35 |
ENST00000374210.6 ENST00000374212.4 |
SLC31A1 |
solute carrier family 31 (copper transporter), member 1 |
| chr5_-_150466692 | 0.34 |
ENST00000315050.7 ENST00000523338.1 ENST00000522100.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr4_+_41614909 | 0.34 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr12_+_113376157 | 0.34 |
ENST00000228928.7 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr19_-_45826125 | 0.33 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr20_+_35807449 | 0.33 |
ENST00000237530.6 |
RPN2 |
ribophorin II |
| chr11_+_108093559 | 0.33 |
ENST00000278616.4 |
ATM |
ataxia telangiectasia mutated |
| chr15_-_83621435 | 0.33 |
ENST00000450735.2 ENST00000426485.1 ENST00000399166.2 ENST00000304231.8 |
HOMER2 |
homer homolog 2 (Drosophila) |
| chr17_-_66287350 | 0.33 |
ENST00000580666.1 ENST00000583477.1 |
SLC16A6 |
solute carrier family 16, member 6 |
| chr8_-_18871159 | 0.32 |
ENST00000327040.8 ENST00000440756.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr17_-_74163159 | 0.32 |
ENST00000591615.1 |
RNF157 |
ring finger protein 157 |
| chr12_+_113376249 | 0.32 |
ENST00000551007.1 ENST00000548514.1 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr17_-_2614927 | 0.32 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr19_-_633576 | 0.32 |
ENST00000588649.2 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
| chr1_+_55464600 | 0.32 |
ENST00000371265.4 |
BSND |
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr16_+_2587998 | 0.31 |
ENST00000441549.3 ENST00000268673.7 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr3_-_9994021 | 0.31 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr16_+_4674787 | 0.31 |
ENST00000262370.7 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr3_+_196439170 | 0.31 |
ENST00000392391.3 ENST00000314118.4 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr2_+_24714729 | 0.31 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr8_-_27462822 | 0.31 |
ENST00000522098.1 |
CLU |
clusterin |
| chr1_+_174769006 | 0.31 |
ENST00000489615.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr4_+_128886424 | 0.31 |
ENST00000398965.1 |
C4orf29 |
chromosome 4 open reading frame 29 |
| chr10_-_118502070 | 0.30 |
ENST00000369209.3 |
HSPA12A |
heat shock 70kDa protein 12A |
| chr17_-_42580738 | 0.30 |
ENST00000585614.1 ENST00000591680.1 ENST00000434000.1 ENST00000588554.1 ENST00000592154.1 |
GPATCH8 |
G patch domain containing 8 |
| chr14_-_24658053 | 0.29 |
ENST00000354464.6 |
IPO4 |
importin 4 |
| chr20_-_48532019 | 0.29 |
ENST00000289431.5 |
SPATA2 |
spermatogenesis associated 2 |
| chr16_+_27413483 | 0.29 |
ENST00000337929.3 ENST00000564089.1 |
IL21R |
interleukin 21 receptor |
| chr1_-_113498616 | 0.29 |
ENST00000433570.4 ENST00000538576.1 ENST00000458229.1 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr17_-_77924627 | 0.29 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr12_-_16759711 | 0.28 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr12_+_121416489 | 0.28 |
ENST00000541395.1 ENST00000544413.1 |
HNF1A |
HNF1 homeobox A |
| chr19_+_7011509 | 0.28 |
ENST00000377296.3 |
AC025278.1 |
Uncharacterized protein |
| chr17_+_39975544 | 0.28 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chrX_-_107975917 | 0.28 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chr7_-_100239132 | 0.28 |
ENST00000223051.3 ENST00000431692.1 |
TFR2 |
transferrin receptor 2 |
| chr9_+_92219919 | 0.27 |
ENST00000252506.6 ENST00000375769.1 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
| chr1_-_33815486 | 0.27 |
ENST00000373418.3 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
| chr19_+_5623186 | 0.27 |
ENST00000538656.1 |
SAFB |
scaffold attachment factor B |
| chr6_-_11807277 | 0.27 |
ENST00000379415.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr16_+_28875126 | 0.27 |
ENST00000359285.5 ENST00000538342.1 |
SH2B1 |
SH2B adaptor protein 1 |
| chr3_+_12330560 | 0.26 |
ENST00000397026.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr17_+_41132564 | 0.26 |
ENST00000361677.1 ENST00000589705.1 |
RUNDC1 |
RUN domain containing 1 |
| chr3_+_44803209 | 0.26 |
ENST00000326047.4 |
KIF15 |
kinesin family member 15 |
| chr7_-_37956409 | 0.26 |
ENST00000436072.2 |
SFRP4 |
secreted frizzled-related protein 4 |
| chr14_-_105262016 | 0.26 |
ENST00000407796.2 |
AKT1 |
v-akt murine thymoma viral oncogene homolog 1 |
| chr2_-_24583583 | 0.25 |
ENST00000355123.4 |
ITSN2 |
intersectin 2 |
| chr6_-_32143828 | 0.25 |
ENST00000412465.2 ENST00000375107.3 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr3_-_125775629 | 0.25 |
ENST00000383598.2 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr7_-_30029367 | 0.25 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chr16_+_4674814 | 0.25 |
ENST00000415496.1 ENST00000587747.1 ENST00000399577.5 ENST00000588994.1 ENST00000586183.1 |
MGRN1 |
mahogunin ring finger 1, E3 ubiquitin protein ligase |
| chr7_+_134528635 | 0.25 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr17_+_79679369 | 0.25 |
ENST00000350690.5 |
SLC25A10 |
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr14_+_75761099 | 0.25 |
ENST00000561000.1 ENST00000558575.1 |
RP11-293M10.5 |
RP11-293M10.5 |
| chr16_+_2587965 | 0.24 |
ENST00000342085.4 ENST00000566659.1 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr2_-_209118974 | 0.24 |
ENST00000415913.1 ENST00000415282.1 ENST00000446179.1 |
IDH1 |
isocitrate dehydrogenase 1 (NADP+), soluble |
| chr3_-_38691119 | 0.24 |
ENST00000333535.4 ENST00000413689.1 ENST00000443581.1 ENST00000425664.1 ENST00000451551.2 |
SCN5A |
sodium channel, voltage-gated, type V, alpha subunit |
| chr2_-_37458749 | 0.24 |
ENST00000234170.5 |
CEBPZ |
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr12_-_16761007 | 0.24 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr6_-_32144838 | 0.23 |
ENST00000395499.1 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr1_+_174933899 | 0.23 |
ENST00000367688.3 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr19_-_4540486 | 0.23 |
ENST00000306390.6 |
LRG1 |
leucine-rich alpha-2-glycoprotein 1 |
| chr6_+_32938665 | 0.23 |
ENST00000374831.4 ENST00000395289.2 |
BRD2 |
bromodomain containing 2 |
| chr1_+_16083154 | 0.23 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr3_+_186435137 | 0.22 |
ENST00000447445.1 |
KNG1 |
kininogen 1 |
| chr11_+_46402297 | 0.22 |
ENST00000405308.2 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr20_+_42839722 | 0.22 |
ENST00000442383.1 ENST00000435163.1 |
OSER1-AS1 |
OSER1 antisense RNA 1 (head to head) |
| chr6_-_34360413 | 0.22 |
ENST00000607016.1 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
| chr11_-_67373435 | 0.22 |
ENST00000446232.1 |
C11orf72 |
chromosome 11 open reading frame 72 |
| chr3_+_10068095 | 0.22 |
ENST00000287647.3 ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2 |
Fanconi anemia, complementation group D2 |
| chr1_+_236849754 | 0.22 |
ENST00000542672.1 ENST00000366578.4 |
ACTN2 |
actinin, alpha 2 |
| chr4_+_41614720 | 0.21 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr4_-_74847800 | 0.21 |
ENST00000296029.3 |
PF4 |
platelet factor 4 |
| chrX_-_40036520 | 0.21 |
ENST00000406200.2 ENST00000378455.4 ENST00000342274.4 |
BCOR |
BCL6 corepressor |
| chr11_+_131781290 | 0.21 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.4 | 2.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.4 | 2.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 2.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 1.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 2.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 2.4 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.7 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 2.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.1 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.4 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) laminin-11 complex(GO:0043260) |
| 0.1 | 0.7 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.1 | 0.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 1.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.2 | GO:0097679 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) other organism cytoplasm(GO:0097679) |
| 0.1 | 0.9 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 2.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 2.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 10.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.2 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 2.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.0 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 1.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.9 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.5 | GO:0055037 | recycling endosome(GO:0055037) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.8 | 4.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.7 | 2.0 | GO:0097327 | response to antineoplastic agent(GO:0097327) |
| 0.6 | 4.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.6 | 1.8 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.6 | 3.0 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.6 | 6.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.5 | 0.9 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.5 | 2.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.4 | 2.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.4 | 1.3 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.4 | 1.9 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.4 | 1.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.3 | 1.0 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.3 | 0.9 | GO:0003285 | septum secundum development(GO:0003285) |
| 0.3 | 0.8 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.3 | 1.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.7 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 1.7 | GO:0031999 | negative regulation of fatty acid beta-oxidation(GO:0031999) |
| 0.2 | 0.7 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.2 | 0.9 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.2 | 0.7 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.2 | 0.7 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.6 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.2 | 0.8 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) phenylpropanoid catabolic process(GO:0046271) |
| 0.2 | 1.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.7 | GO:1990535 | transformation of host cell by virus(GO:0019087) neuron projection maintenance(GO:1990535) |
| 0.2 | 0.7 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 0.2 | 0.5 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) |
| 0.2 | 0.3 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.2 | 0.6 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 0.6 | GO:0006064 | xylulose metabolic process(GO:0005997) glucuronate catabolic process(GO:0006064) glucuronate catabolic process to xylulose 5-phosphate(GO:0019640) xylulose 5-phosphate metabolic process(GO:0051167) xylulose 5-phosphate biosynthetic process(GO:1901159) |
| 0.1 | 1.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.1 | 2.6 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.1 | 0.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.1 | 0.9 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 1.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.4 | GO:0015743 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 1.4 | GO:0034370 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) |
| 0.1 | 1.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.3 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 1.0 | GO:0071313 | cellular response to caffeine(GO:0071313) |
| 0.1 | 0.3 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.9 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.3 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.5 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 1.0 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.7 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 1.6 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.5 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.4 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.9 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.3 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.2 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.1 | 0.9 | GO:2000291 | regulation of myoblast proliferation(GO:2000291) |
| 0.1 | 0.6 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.3 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.7 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.2 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.4 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.1 | 1.0 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.1 | 0.7 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.1 | 0.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 2.5 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.1 | 0.6 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.3 | GO:1902998 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.8 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) |
| 0.0 | 0.2 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0060266 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.6 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.4 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.2 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.6 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.3 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0072366 | regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.4 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.5 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.3 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 1.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.0 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.1 | GO:2000317 | T-helper 1 cell lineage commitment(GO:0002296) cellular response to mercury ion(GO:0071288) negative regulation of complement-dependent cytotoxicity(GO:1903660) negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.9 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 1.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.6 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.0 | 0.4 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.6 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.8 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.0 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0019918 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:1903593 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0061008 | liver development(GO:0001889) hepaticobiliary system development(GO:0061008) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.0 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 1.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 5.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 1.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.9 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.8 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 2.6 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.9 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 1.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.9 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.1 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 1.3 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.7 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 8.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 2.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.3 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.9 | 3.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.5 | 2.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 1.9 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.4 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 1.8 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.4 | 2.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 2.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.3 | 1.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.3 | 0.8 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.2 | 1.5 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.2 | 0.7 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.2 | 1.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.6 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.2 | 1.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 1.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.0 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.9 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 0.7 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 1.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 1.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.4 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 1.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 2.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.1 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.1 | 0.7 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 0.4 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 2.0 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.7 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.7 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.1 | 0.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.7 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.6 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 5.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.2 | GO:0052848 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 0.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.6 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 2.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) diacylglycerol binding(GO:0019992) |
| 0.0 | 1.5 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.8 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.5 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.3 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.1 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.8 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.0 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |