Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for OLIG1
Z-value: 0.58
Transcription factors associated with OLIG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
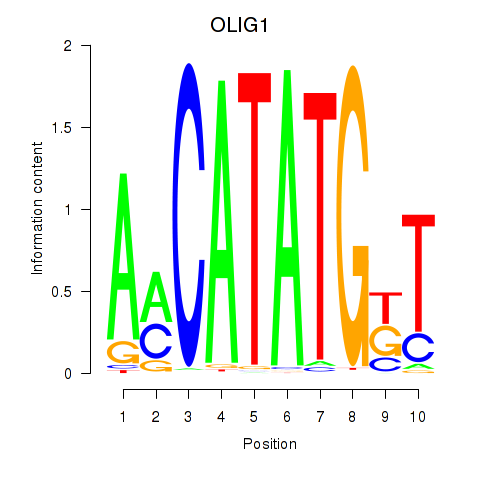
OLIG1
|
ENSG00000184221.8 | OLIG1 |
Activity profile of OLIG1 motif
Sorted Z-values of OLIG1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_158345462 | 1.42 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr3_-_121379739 | 1.24 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr7_-_55930443 | 0.77 |
ENST00000388975.3 |
SEPT14 |
septin 14 |
| chr1_+_220863187 | 0.76 |
ENST00000294889.5 |
C1orf115 |
chromosome 1 open reading frame 115 |
| chr13_-_46679185 | 0.76 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr3_-_148939835 | 0.75 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr14_+_95047725 | 0.73 |
ENST00000554760.1 ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr16_+_12059091 | 0.70 |
ENST00000562385.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr2_+_201994042 | 0.69 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr14_+_95047744 | 0.67 |
ENST00000553511.1 ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr2_+_201994208 | 0.64 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr16_+_12059050 | 0.62 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr16_+_12058961 | 0.61 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr15_-_45670924 | 0.60 |
ENST00000396659.3 |
GATM |
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr2_+_11674213 | 0.59 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr2_+_177053307 | 0.59 |
ENST00000331462.4 |
HOXD1 |
homeobox D1 |
| chr17_-_7017559 | 0.57 |
ENST00000446679.2 |
ASGR2 |
asialoglycoprotein receptor 2 |
| chr4_+_74301880 | 0.55 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr3_-_98241358 | 0.51 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr19_+_18208603 | 0.50 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr3_-_58200398 | 0.47 |
ENST00000318316.3 ENST00000460422.1 ENST00000483681.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr1_-_231560790 | 0.47 |
ENST00000366641.3 |
EGLN1 |
egl-9 family hypoxia-inducible factor 1 |
| chr3_+_113251143 | 0.43 |
ENST00000264852.4 ENST00000393830.3 |
SIDT1 |
SID1 transmembrane family, member 1 |
| chr2_-_166930131 | 0.42 |
ENST00000303395.4 ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A |
sodium channel, voltage-gated, type I, alpha subunit |
| chr7_-_37024665 | 0.40 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chr6_+_292051 | 0.40 |
ENST00000344450.5 |
DUSP22 |
dual specificity phosphatase 22 |
| chr8_-_95449155 | 0.40 |
ENST00000481490.2 |
FSBP |
fibrinogen silencer binding protein |
| chrX_+_69509927 | 0.40 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr21_+_42792442 | 0.39 |
ENST00000398600.2 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr10_-_11574274 | 0.39 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr2_+_201994569 | 0.39 |
ENST00000457277.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr2_-_165424973 | 0.39 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr11_+_65405556 | 0.38 |
ENST00000534313.1 ENST00000533361.1 ENST00000526137.1 |
SIPA1 |
signal-induced proliferation-associated 1 |
| chr14_+_21249200 | 0.38 |
ENST00000304677.2 |
RNASE6 |
ribonuclease, RNase A family, k6 |
| chr16_-_55867146 | 0.38 |
ENST00000422046.2 |
CES1 |
carboxylesterase 1 |
| chrX_+_105855160 | 0.37 |
ENST00000372544.2 ENST00000372548.4 |
CXorf57 |
chromosome X open reading frame 57 |
| chr5_-_150460539 | 0.36 |
ENST00000520931.1 ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr4_+_75858318 | 0.36 |
ENST00000307428.7 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr7_+_4721885 | 0.35 |
ENST00000328914.4 |
FOXK1 |
forkhead box K1 |
| chr2_-_183106641 | 0.31 |
ENST00000346717.4 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr10_-_97416400 | 0.31 |
ENST00000371224.2 ENST00000371221.3 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
| chr14_-_55658252 | 0.31 |
ENST00000395425.2 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
| chr13_-_99910673 | 0.30 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr1_+_244214577 | 0.30 |
ENST00000358704.4 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
| chr5_-_169725231 | 0.30 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr1_+_203274639 | 0.30 |
ENST00000290551.4 |
BTG2 |
BTG family, member 2 |
| chr3_-_197300194 | 0.30 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr10_-_69597915 | 0.30 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr4_+_75858290 | 0.29 |
ENST00000513238.1 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr4_+_146402925 | 0.29 |
ENST00000302085.4 |
SMAD1 |
SMAD family member 1 |
| chr14_-_55658323 | 0.28 |
ENST00000554067.1 ENST00000247191.2 |
DLGAP5 |
discs, large (Drosophila) homolog-associated protein 5 |
| chr19_+_18496957 | 0.28 |
ENST00000252809.3 |
GDF15 |
growth differentiation factor 15 |
| chr4_-_153456153 | 0.28 |
ENST00000603548.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr14_+_88471468 | 0.28 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr7_-_11871815 | 0.28 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr4_-_159094194 | 0.28 |
ENST00000592057.1 ENST00000585682.1 ENST00000393807.5 |
FAM198B |
family with sequence similarity 198, member B |
| chr4_+_160188889 | 0.27 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr10_-_69597810 | 0.27 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr13_+_49551020 | 0.27 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr1_-_160549235 | 0.26 |
ENST00000368054.3 ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84 |
CD84 molecule |
| chr2_-_175462934 | 0.26 |
ENST00000392546.2 ENST00000436221.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr4_+_69313145 | 0.25 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr17_+_41003166 | 0.25 |
ENST00000308423.2 |
AOC3 |
amine oxidase, copper containing 3 |
| chr1_+_50569575 | 0.24 |
ENST00000371827.1 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr5_+_59726565 | 0.24 |
ENST00000412930.2 |
FKSG52 |
FKSG52 |
| chr12_-_102872317 | 0.23 |
ENST00000424202.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr22_+_24999114 | 0.22 |
ENST00000412658.1 ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr4_-_74088800 | 0.22 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr7_-_99573640 | 0.21 |
ENST00000411734.1 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr22_-_19435755 | 0.21 |
ENST00000542103.1 ENST00000399562.4 |
C22orf39 |
chromosome 22 open reading frame 39 |
| chr12_-_40013553 | 0.21 |
ENST00000308666.3 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr6_-_160147925 | 0.21 |
ENST00000535561.1 |
SOD2 |
superoxide dismutase 2, mitochondrial |
| chrX_+_107288197 | 0.21 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr10_+_26727125 | 0.21 |
ENST00000376236.4 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr8_+_86121448 | 0.20 |
ENST00000520225.1 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr19_-_52227221 | 0.20 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr8_+_21915368 | 0.20 |
ENST00000265800.5 ENST00000517418.1 |
DMTN |
dematin actin binding protein |
| chr3_+_171758344 | 0.20 |
ENST00000336824.4 ENST00000423424.1 |
FNDC3B |
fibronectin type III domain containing 3B |
| chrX_-_154563889 | 0.18 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr3_-_33700933 | 0.18 |
ENST00000480013.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr2_+_54342574 | 0.17 |
ENST00000303536.4 ENST00000394666.3 |
ACYP2 |
acylphosphatase 2, muscle type |
| chr19_+_45542773 | 0.17 |
ENST00000544944.2 |
CLASRP |
CLK4-associating serine/arginine rich protein |
| chr15_-_38519066 | 0.17 |
ENST00000561320.1 ENST00000561161.1 |
RP11-346D14.1 |
RP11-346D14.1 |
| chr3_-_178984759 | 0.17 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chrX_+_107288239 | 0.17 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr14_-_69864993 | 0.17 |
ENST00000555373.1 |
ERH |
enhancer of rudimentary homolog (Drosophila) |
| chr7_-_27205136 | 0.16 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr5_-_115872142 | 0.16 |
ENST00000510263.1 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr12_-_108714412 | 0.16 |
ENST00000412676.1 ENST00000550573.1 |
CMKLR1 |
chemokine-like receptor 1 |
| chr2_+_191792376 | 0.16 |
ENST00000409428.1 ENST00000409215.1 |
GLS |
glutaminase |
| chr1_+_57320437 | 0.15 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr6_+_135502501 | 0.15 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_-_41216619 | 0.15 |
ENST00000508676.1 ENST00000506352.1 ENST00000295974.8 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr2_+_241807870 | 0.15 |
ENST00000307503.3 |
AGXT |
alanine-glyoxylate aminotransferase |
| chr11_+_92085707 | 0.15 |
ENST00000525166.1 |
FAT3 |
FAT atypical cadherin 3 |
| chr4_+_2470664 | 0.14 |
ENST00000314289.8 ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4 |
ring finger protein 4 |
| chr16_+_14726672 | 0.14 |
ENST00000261658.2 ENST00000563971.1 |
BFAR |
bifunctional apoptosis regulator |
| chr1_+_169764163 | 0.14 |
ENST00000413811.2 ENST00000359326.4 ENST00000456684.1 |
C1orf112 |
chromosome 1 open reading frame 112 |
| chr6_+_10528560 | 0.14 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr8_-_38239732 | 0.14 |
ENST00000534155.1 ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr5_-_131132658 | 0.14 |
ENST00000514667.1 ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3 FNIP1 |
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr3_+_122103014 | 0.13 |
ENST00000232125.5 ENST00000477892.1 ENST00000469967.1 |
FAM162A |
family with sequence similarity 162, member A |
| chr1_+_91966384 | 0.13 |
ENST00000430031.2 ENST00000234626.6 |
CDC7 |
cell division cycle 7 |
| chr1_+_110881945 | 0.13 |
ENST00000602849.1 ENST00000487146.2 |
RBM15 |
RNA binding motif protein 15 |
| chr10_-_94257512 | 0.13 |
ENST00000371581.5 |
IDE |
insulin-degrading enzyme |
| chr3_+_119298429 | 0.13 |
ENST00000478927.1 |
ADPRH |
ADP-ribosylarginine hydrolase |
| chr1_+_91966656 | 0.13 |
ENST00000428239.1 ENST00000426137.1 |
CDC7 |
cell division cycle 7 |
| chr5_-_11588907 | 0.12 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr4_-_120243545 | 0.12 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr19_-_39368887 | 0.12 |
ENST00000340740.3 ENST00000591812.1 |
RINL |
Ras and Rab interactor-like |
| chr5_+_157158205 | 0.11 |
ENST00000231198.7 |
THG1L |
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr18_-_47813940 | 0.11 |
ENST00000586837.1 ENST00000412036.2 ENST00000589940.1 |
CXXC1 |
CXXC finger protein 1 |
| chr4_-_174451370 | 0.11 |
ENST00000359562.4 |
HAND2 |
heart and neural crest derivatives expressed 2 |
| chr3_-_195310802 | 0.11 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr3_-_101232019 | 0.11 |
ENST00000394095.2 ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7 |
SUMO1/sentrin specific peptidase 7 |
| chr11_-_111741994 | 0.11 |
ENST00000398006.2 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
| chr6_+_37400974 | 0.10 |
ENST00000455891.1 ENST00000373451.4 |
CMTR1 |
cap methyltransferase 1 |
| chr22_+_36113919 | 0.10 |
ENST00000249044.2 |
APOL5 |
apolipoprotein L, 5 |
| chr12_-_16761007 | 0.10 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr2_-_183387064 | 0.10 |
ENST00000536095.1 ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr12_+_56618102 | 0.09 |
ENST00000267023.4 ENST00000380198.2 ENST00000341463.5 |
NABP2 |
nucleic acid binding protein 2 |
| chr2_-_183387283 | 0.09 |
ENST00000435564.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr19_+_12035913 | 0.09 |
ENST00000591944.1 |
ZNF763 |
Uncharacterized protein; Zinc finger protein 763 |
| chr3_-_114477787 | 0.09 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_-_145701718 | 0.09 |
ENST00000377317.4 |
FOXH1 |
forkhead box H1 |
| chr1_+_76251879 | 0.09 |
ENST00000535300.1 ENST00000319942.3 |
RABGGTB |
Rab geranylgeranyltransferase, beta subunit |
| chr1_+_24969755 | 0.09 |
ENST00000447431.2 ENST00000374389.4 |
SRRM1 |
serine/arginine repetitive matrix 1 |
| chr1_-_205091115 | 0.09 |
ENST00000264515.6 ENST00000367164.1 |
RBBP5 |
retinoblastoma binding protein 5 |
| chr1_+_15479054 | 0.08 |
ENST00000376014.3 ENST00000451326.2 |
TMEM51 |
transmembrane protein 51 |
| chr4_+_118955500 | 0.08 |
ENST00000296499.5 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr2_-_228582709 | 0.08 |
ENST00000541617.1 ENST00000409456.2 ENST00000409287.1 ENST00000258403.3 |
SLC19A3 |
solute carrier family 19 (thiamine transporter), member 3 |
| chr2_-_152830441 | 0.08 |
ENST00000534999.1 ENST00000397327.2 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr7_-_38403077 | 0.08 |
ENST00000426402.2 |
TRGV2 |
T cell receptor gamma variable 2 |
| chr1_+_146714291 | 0.08 |
ENST00000431239.1 ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
| chr6_-_31648150 | 0.08 |
ENST00000375858.3 ENST00000383237.4 |
LY6G5C |
lymphocyte antigen 6 complex, locus G5C |
| chr5_-_75919253 | 0.08 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr1_+_15479021 | 0.08 |
ENST00000428417.1 |
TMEM51 |
transmembrane protein 51 |
| chr5_-_11589131 | 0.08 |
ENST00000511377.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr10_+_51565108 | 0.08 |
ENST00000438493.1 ENST00000452682.1 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr11_-_107582775 | 0.08 |
ENST00000305991.2 |
SLN |
sarcolipin |
| chr9_-_19786926 | 0.08 |
ENST00000341998.2 ENST00000286344.3 |
SLC24A2 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr20_+_48429233 | 0.07 |
ENST00000417961.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr10_+_51565188 | 0.07 |
ENST00000430396.2 ENST00000374087.4 ENST00000414907.2 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr7_+_64838786 | 0.07 |
ENST00000450302.2 |
ZNF92 |
zinc finger protein 92 |
| chr20_+_57226841 | 0.07 |
ENST00000358029.4 ENST00000361830.3 |
STX16 |
syntaxin 16 |
| chr1_+_153750622 | 0.07 |
ENST00000532853.1 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr1_+_180897269 | 0.07 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chr16_+_72142195 | 0.07 |
ENST00000563819.1 ENST00000567142.2 |
DHX38 |
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr6_+_163148161 | 0.07 |
ENST00000337019.3 ENST00000366889.2 |
PACRG |
PARK2 co-regulated |
| chr3_-_33700589 | 0.07 |
ENST00000461133.3 ENST00000496954.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr18_+_32173276 | 0.07 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chrX_-_111326004 | 0.06 |
ENST00000262839.2 |
TRPC5 |
transient receptor potential cation channel, subfamily C, member 5 |
| chr8_+_119294456 | 0.06 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chr2_+_75873902 | 0.06 |
ENST00000393909.2 ENST00000358788.6 ENST00000409374.1 |
MRPL19 |
mitochondrial ribosomal protein L19 |
| chr6_-_46922659 | 0.06 |
ENST00000265417.7 |
GPR116 |
G protein-coupled receptor 116 |
| chr3_-_49466686 | 0.06 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr3_-_114477962 | 0.06 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr19_-_51472222 | 0.06 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr12_-_123450986 | 0.06 |
ENST00000344275.7 ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr12_+_26126681 | 0.06 |
ENST00000542865.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr12_+_133657461 | 0.06 |
ENST00000412146.2 ENST00000544426.1 ENST00000440984.2 ENST00000319849.3 ENST00000440550.2 |
ZNF140 |
zinc finger protein 140 |
| chr1_-_169396666 | 0.06 |
ENST00000456107.1 ENST00000367805.3 |
CCDC181 |
coiled-coil domain containing 181 |
| chr6_+_52051171 | 0.06 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chr5_-_75919217 | 0.06 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr11_-_88796803 | 0.06 |
ENST00000418177.2 ENST00000455756.2 |
GRM5 |
glutamate receptor, metabotropic 5 |
| chr16_-_71843047 | 0.05 |
ENST00000299980.4 ENST00000393512.3 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
| chr5_-_137674000 | 0.05 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr4_-_76912070 | 0.05 |
ENST00000395711.4 ENST00000356260.5 |
SDAD1 |
SDA1 domain containing 1 |
| chr1_-_101491319 | 0.05 |
ENST00000342173.7 ENST00000488176.1 ENST00000370109.3 |
DPH5 |
diphthamide biosynthesis 5 |
| chr4_-_70505358 | 0.05 |
ENST00000457664.2 ENST00000604629.1 ENST00000604021.1 |
UGT2A2 |
UDP glucuronosyltransferase 2 family, polypeptide A2 |
| chr1_-_169396646 | 0.05 |
ENST00000367806.3 |
CCDC181 |
coiled-coil domain containing 181 |
| chr1_-_92951607 | 0.05 |
ENST00000427103.1 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr3_-_46068969 | 0.05 |
ENST00000542109.1 ENST00000395946.2 |
XCR1 |
chemokine (C motif) receptor 1 |
| chr11_-_44972418 | 0.05 |
ENST00000525680.1 ENST00000528290.1 ENST00000530035.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr19_-_11450249 | 0.05 |
ENST00000222120.3 |
RAB3D |
RAB3D, member RAS oncogene family |
| chr4_-_83280767 | 0.05 |
ENST00000514671.1 |
HNRNPD |
heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) |
| chr2_-_40006357 | 0.05 |
ENST00000505747.1 |
THUMPD2 |
THUMP domain containing 2 |
| chr20_-_44144249 | 0.05 |
ENST00000217428.6 |
SPINT3 |
serine peptidase inhibitor, Kunitz type, 3 |
| chr2_-_70418032 | 0.04 |
ENST00000425268.1 ENST00000428751.1 ENST00000417203.1 ENST00000417865.1 ENST00000428010.1 ENST00000447804.1 ENST00000264434.2 |
C2orf42 |
chromosome 2 open reading frame 42 |
| chr1_-_111970353 | 0.04 |
ENST00000369732.3 |
OVGP1 |
oviductal glycoprotein 1, 120kDa |
| chr6_-_25874440 | 0.04 |
ENST00000361703.6 ENST00000397060.4 |
SLC17A3 |
solute carrier family 17 (organic anion transporter), member 3 |
| chr1_+_156182773 | 0.04 |
ENST00000490491.1 ENST00000368276.4 ENST00000320139.5 ENST00000368279.3 ENST00000368273.4 ENST00000368277.3 ENST00000567140.1 ENST00000565805.1 |
PMF1-BGLAP PMF1 |
PMF1-BGLAP readthrough polyamine-modulated factor 1 |
| chr1_+_196857144 | 0.04 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr19_+_21324827 | 0.04 |
ENST00000600692.1 ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431 |
zinc finger protein 431 |
| chr11_-_11374904 | 0.04 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
| chr15_-_65503801 | 0.04 |
ENST00000261883.4 |
CILP |
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr16_-_71842941 | 0.04 |
ENST00000423132.2 ENST00000433195.2 ENST00000569748.1 ENST00000570017.1 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
| chr15_-_80189380 | 0.03 |
ENST00000258874.3 |
MTHFS |
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr4_+_154178520 | 0.03 |
ENST00000433687.1 |
TRIM2 |
tripartite motif containing 2 |
| chr6_-_28891709 | 0.03 |
ENST00000377194.3 ENST00000377199.3 |
TRIM27 |
tripartite motif containing 27 |
| chr15_-_50557863 | 0.03 |
ENST00000543581.1 |
HDC |
histidine decarboxylase |
| chr11_+_89764274 | 0.03 |
ENST00000448984.1 ENST00000432771.1 |
TRIM49C |
tripartite motif containing 49C |
| chrX_+_118602363 | 0.03 |
ENST00000317881.8 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr5_+_118812237 | 0.03 |
ENST00000513628.1 |
HSD17B4 |
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr1_+_100810575 | 0.02 |
ENST00000542213.1 |
CDC14A |
cell division cycle 14A |
| chrX_+_118108601 | 0.02 |
ENST00000371628.3 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chr4_+_144354644 | 0.02 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr17_+_34842473 | 0.02 |
ENST00000490126.2 ENST00000225410.4 |
ZNHIT3 |
zinc finger, HIT-type containing 3 |
| chr3_-_46069223 | 0.02 |
ENST00000309285.3 |
XCR1 |
chemokine (C motif) receptor 1 |
| chr1_+_104198377 | 0.02 |
ENST00000370083.4 |
AMY1A |
amylase, alpha 1A (salivary) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0036029 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 1.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.4 | GO:0043194 | voltage-gated sodium channel complex(GO:0001518) axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.1 | 0.6 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.4 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 1.7 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.3 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.7 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.2 | 0.8 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 1.7 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.6 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.3 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.5 | GO:0030805 | regulation of cyclic nucleotide catabolic process(GO:0030805) regulation of cAMP catabolic process(GO:0030820) regulation of purine nucleotide catabolic process(GO:0033121) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.1 | 1.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.6 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:0006524 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.2 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.2 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.4 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.4 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.5 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.4 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.2 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.2 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.5 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.0 | 0.2 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.1 | GO:0071934 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 1.9 | GO:0002260 | lymphocyte homeostasis(GO:0002260) |
| 0.0 | 0.1 | GO:1901877 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.2 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.0 | 0.1 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.2 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.1 | GO:0061032 | cardiac right ventricle formation(GO:0003219) visceral serous pericardium development(GO:0061032) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.3 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.3 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.0 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.0 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.2 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0032667 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |