Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
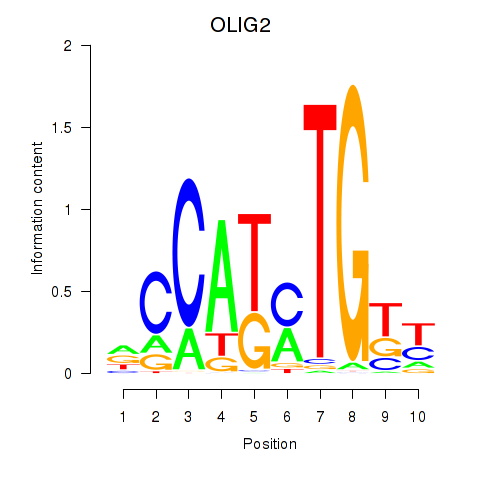
Results for OLIG2_NEUROD1_ATOH1
Z-value: 1.21
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.4 | OLIG2 |
|
NEUROD1
|
ENSG00000162992.3 | NEUROD1 |
|
ATOH1
|
ENSG00000172238.3 | ATOH1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OLIG2 | hg19_v2_chr21_+_34398153_34398250 | 0.20 | 4.6e-01 | Click! |
| NEUROD1 | hg19_v2_chr2_-_182545603_182545603 | 0.20 | 4.7e-01 | Click! |
| ATOH1 | hg19_v2_chr4_+_94750014_94750042 | -0.04 | 8.8e-01 | Click! |
Activity profile of OLIG2_NEUROD1_ATOH1 motif
Sorted Z-values of OLIG2_NEUROD1_ATOH1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG2_NEUROD1_ATOH1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_49834299 | 4.28 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr16_+_30386098 | 4.13 |
ENST00000322861.7 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr3_-_52486841 | 4.03 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr8_-_49833978 | 4.02 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr12_+_6309963 | 3.97 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr5_+_135364584 | 3.41 |
ENST00000442011.2 ENST00000305126.8 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr12_+_6309517 | 3.39 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chrX_+_135278908 | 3.32 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr5_+_102201722 | 3.17 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr3_-_99833333 | 3.03 |
ENST00000354552.3 ENST00000331335.5 ENST00000398326.2 |
FILIP1L |
filamin A interacting protein 1-like |
| chrX_+_135279179 | 3.00 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr2_-_163100045 | 3.00 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr14_-_75079026 | 2.98 |
ENST00000261978.4 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
| chr5_+_102201509 | 2.96 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr2_-_152590946 | 2.92 |
ENST00000172853.10 |
NEB |
nebulin |
| chr15_+_63354769 | 2.91 |
ENST00000558910.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr16_+_30383613 | 2.87 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr21_+_30502806 | 2.76 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr6_+_44184653 | 2.49 |
ENST00000573382.2 ENST00000576476.1 |
RP1-302G2.5 |
RP1-302G2.5 |
| chr3_+_159570722 | 2.39 |
ENST00000482804.1 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr13_-_33780133 | 2.38 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr1_-_201346761 | 2.33 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr13_+_102104980 | 2.22 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_-_16936340 | 2.13 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr13_+_102104952 | 2.10 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_+_102201687 | 1.84 |
ENST00000304400.7 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr11_+_32112431 | 1.80 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr5_+_102201430 | 1.78 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr8_+_70378852 | 1.75 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr9_+_116298778 | 1.62 |
ENST00000462143.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr2_-_175711133 | 1.54 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr12_+_53491220 | 1.54 |
ENST00000548547.1 ENST00000301464.3 |
IGFBP6 |
insulin-like growth factor binding protein 6 |
| chr11_+_117073850 | 1.49 |
ENST00000529622.1 |
TAGLN |
transgelin |
| chr11_+_44587141 | 1.41 |
ENST00000227155.4 ENST00000342935.3 ENST00000532544.1 |
CD82 |
CD82 molecule |
| chr7_-_30029574 | 1.41 |
ENST00000426154.1 ENST00000421434.1 ENST00000434476.2 |
SCRN1 |
secernin 1 |
| chr11_-_2170786 | 1.39 |
ENST00000300632.5 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr9_-_35689900 | 1.31 |
ENST00000378300.5 ENST00000329305.2 ENST00000360958.2 |
TPM2 |
tropomyosin 2 (beta) |
| chr7_-_30029367 | 1.27 |
ENST00000242059.5 |
SCRN1 |
secernin 1 |
| chrX_+_70521584 | 1.23 |
ENST00000373829.3 ENST00000538820.1 |
ITGB1BP2 |
integrin beta 1 binding protein (melusin) 2 |
| chr7_+_73442422 | 1.22 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chr7_-_151511911 | 1.20 |
ENST00000392801.2 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr12_-_123752624 | 1.16 |
ENST00000542174.1 ENST00000535796.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr7_+_143079000 | 1.15 |
ENST00000392910.2 |
ZYX |
zyxin |
| chr19_-_45826125 | 1.14 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr7_+_73442487 | 1.14 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr1_-_193155729 | 1.14 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr12_+_49212514 | 1.12 |
ENST00000301050.2 ENST00000548279.1 ENST00000547230.1 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
| chr20_-_56284816 | 1.11 |
ENST00000395819.3 ENST00000341744.3 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr17_-_15168624 | 1.10 |
ENST00000312280.3 ENST00000494511.1 ENST00000580584.1 |
PMP22 |
peripheral myelin protein 22 |
| chr22_+_45072958 | 1.09 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr1_+_172422026 | 1.08 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr1_+_114522049 | 1.07 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr1_-_153522562 | 1.06 |
ENST00000368714.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr22_+_45072925 | 1.05 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr3_-_87040233 | 1.01 |
ENST00000398399.2 |
VGLL3 |
vestigial like 3 (Drosophila) |
| chrX_+_47444613 | 0.96 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr7_+_143078652 | 0.95 |
ENST00000354434.4 ENST00000449423.2 |
ZYX |
zyxin |
| chr22_-_36236623 | 0.94 |
ENST00000405409.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr9_+_92219919 | 0.93 |
ENST00000252506.6 ENST00000375769.1 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
| chr15_+_63340553 | 0.92 |
ENST00000334895.5 |
TPM1 |
tropomyosin 1 (alpha) |
| chr3_-_18480260 | 0.92 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr7_+_73442457 | 0.91 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chr19_-_46285646 | 0.91 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr5_-_111312622 | 0.90 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chrX_-_10544942 | 0.90 |
ENST00000380779.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr22_-_36236265 | 0.87 |
ENST00000414461.2 ENST00000416721.2 ENST00000449924.2 ENST00000262829.7 ENST00000397305.3 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr6_-_24911195 | 0.87 |
ENST00000259698.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr12_+_122150646 | 0.85 |
ENST00000449592.2 |
TMEM120B |
transmembrane protein 120B |
| chr17_+_60758814 | 0.81 |
ENST00000579432.1 ENST00000446119.2 |
MRC2 |
mannose receptor, C type 2 |
| chr19_+_16186903 | 0.81 |
ENST00000588507.1 |
TPM4 |
tropomyosin 4 |
| chrX_-_10645773 | 0.80 |
ENST00000453318.2 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr19_+_16187085 | 0.79 |
ENST00000300933.4 |
TPM4 |
tropomyosin 4 |
| chr19_-_46285736 | 0.78 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chr3_+_8775466 | 0.78 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chrX_+_54835493 | 0.76 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr8_+_26371763 | 0.75 |
ENST00000521913.1 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chrX_-_13835147 | 0.74 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr6_-_52860171 | 0.74 |
ENST00000370963.4 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr15_-_63448973 | 0.72 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr16_+_6533729 | 0.72 |
ENST00000551752.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr19_-_19051103 | 0.70 |
ENST00000542541.2 ENST00000433218.2 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr5_+_34656569 | 0.70 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr7_-_41742697 | 0.69 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr5_+_148521381 | 0.68 |
ENST00000504238.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr1_+_25071848 | 0.68 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr2_-_179672142 | 0.67 |
ENST00000342992.6 ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN |
titin |
| chr7_+_75931861 | 0.66 |
ENST00000248553.6 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr14_-_53417732 | 0.66 |
ENST00000399304.3 ENST00000395631.2 ENST00000341590.3 ENST00000343279.4 |
FERMT2 |
fermitin family member 2 |
| chr5_+_148521046 | 0.66 |
ENST00000326685.7 ENST00000356541.3 ENST00000309868.7 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr6_+_129204337 | 0.65 |
ENST00000421865.2 |
LAMA2 |
laminin, alpha 2 |
| chr13_+_38923959 | 0.65 |
ENST00000379649.1 ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1 |
ubiquitin-fold modifier 1 |
| chr5_+_148521136 | 0.65 |
ENST00000506113.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr12_-_49318715 | 0.64 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr8_-_145018905 | 0.64 |
ENST00000398774.2 |
PLEC |
plectin |
| chr11_-_66084508 | 0.64 |
ENST00000311330.3 |
CD248 |
CD248 molecule, endosialin |
| chr17_-_37009882 | 0.64 |
ENST00000378096.3 ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23 |
ribosomal protein L23 |
| chr13_-_36705425 | 0.63 |
ENST00000255448.4 ENST00000360631.3 ENST00000379892.4 |
DCLK1 |
doublecortin-like kinase 1 |
| chr12_+_26348246 | 0.63 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr12_+_6833237 | 0.63 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr7_+_73442102 | 0.61 |
ENST00000445912.1 ENST00000252034.7 |
ELN |
elastin |
| chr11_+_46299199 | 0.61 |
ENST00000529193.1 ENST00000288400.3 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr1_-_17304771 | 0.61 |
ENST00000375534.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr12_+_6833437 | 0.59 |
ENST00000534947.1 ENST00000541866.1 ENST00000534877.1 ENST00000538753.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr22_+_31488433 | 0.58 |
ENST00000455608.1 |
SMTN |
smoothelin |
| chr12_+_26348429 | 0.58 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr9_-_14313641 | 0.58 |
ENST00000380953.1 |
NFIB |
nuclear factor I/B |
| chrX_+_154611749 | 0.56 |
ENST00000369505.3 |
F8A2 |
coagulation factor VIII-associated 2 |
| chr9_-_14313893 | 0.56 |
ENST00000380921.3 ENST00000380959.3 |
NFIB |
nuclear factor I/B |
| chr14_+_90863327 | 0.56 |
ENST00000356978.4 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr15_-_42749711 | 0.54 |
ENST00000565611.1 ENST00000263805.4 ENST00000565948.1 |
ZNF106 |
zinc finger protein 106 |
| chr3_-_45267760 | 0.54 |
ENST00000503771.1 |
TMEM158 |
transmembrane protein 158 (gene/pseudogene) |
| chr21_-_39288743 | 0.54 |
ENST00000609713.1 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr6_-_112194484 | 0.53 |
ENST00000518295.1 ENST00000484067.2 ENST00000229470.5 ENST00000356013.2 ENST00000368678.4 ENST00000523238.1 ENST00000354650.3 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr12_-_7245125 | 0.53 |
ENST00000542285.1 ENST00000540610.1 |
C1R |
complement component 1, r subcomponent |
| chr4_-_186877806 | 0.52 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr4_-_41216492 | 0.52 |
ENST00000503503.1 ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chrX_-_106243451 | 0.51 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr17_+_48243352 | 0.51 |
ENST00000344627.6 ENST00000262018.3 ENST00000543315.1 ENST00000451235.2 ENST00000511303.1 |
SGCA |
sarcoglycan, alpha (50kDa dystrophin-associated glycoprotein) |
| chr1_-_161993422 | 0.49 |
ENST00000367940.2 |
OLFML2B |
olfactomedin-like 2B |
| chr12_-_49319265 | 0.49 |
ENST00000552878.1 ENST00000453172.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr16_+_6533380 | 0.47 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr17_-_46623441 | 0.47 |
ENST00000330070.4 |
HOXB2 |
homeobox B2 |
| chr14_+_90863364 | 0.47 |
ENST00000447653.3 ENST00000553542.1 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
| chr1_-_33168336 | 0.47 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr4_-_41216473 | 0.47 |
ENST00000513140.1 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr1_+_180165672 | 0.46 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr12_-_52845910 | 0.46 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr4_-_186877502 | 0.44 |
ENST00000431902.1 ENST00000284776.7 ENST00000415274.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr9_-_13165457 | 0.44 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr15_-_73925651 | 0.44 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr4_-_103749105 | 0.44 |
ENST00000394801.4 ENST00000394804.2 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_-_52002403 | 0.44 |
ENST00000490063.1 ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4 |
poly(rC) binding protein 4 |
| chr3_-_149093499 | 0.44 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr8_-_145018080 | 0.43 |
ENST00000354589.3 |
PLEC |
plectin |
| chr22_+_31489344 | 0.43 |
ENST00000404574.1 |
SMTN |
smoothelin |
| chr4_-_41216619 | 0.43 |
ENST00000508676.1 ENST00000506352.1 ENST00000295974.8 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr9_-_35111420 | 0.43 |
ENST00000378557.1 |
FAM214B |
family with sequence similarity 214, member B |
| chr5_-_171433579 | 0.43 |
ENST00000265094.5 ENST00000393802.2 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr10_-_15413035 | 0.42 |
ENST00000378116.4 ENST00000455654.1 |
FAM171A1 |
family with sequence similarity 171, member A1 |
| chrX_+_152082969 | 0.42 |
ENST00000535861.1 ENST00000539731.1 ENST00000449285.2 ENST00000318504.7 ENST00000324823.6 ENST00000370268.4 ENST00000370270.2 |
ZNF185 |
zinc finger protein 185 (LIM domain) |
| chr10_+_96162242 | 0.42 |
ENST00000225235.4 |
TBC1D12 |
TBC1 domain family, member 12 |
| chr17_-_3595181 | 0.42 |
ENST00000552050.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr11_-_2182388 | 0.42 |
ENST00000421783.1 ENST00000397262.1 ENST00000250971.3 ENST00000381330.4 ENST00000397270.1 |
INS INS-IGF2 |
insulin INS-IGF2 readthrough |
| chr12_+_4382917 | 0.41 |
ENST00000261254.3 |
CCND2 |
cyclin D2 |
| chr6_-_52859968 | 0.40 |
ENST00000370959.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr15_+_43809797 | 0.40 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr8_+_54764346 | 0.40 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr3_+_46919235 | 0.39 |
ENST00000449590.1 |
PTH1R |
parathyroid hormone 1 receptor |
| chr17_+_65040678 | 0.39 |
ENST00000226021.3 |
CACNG1 |
calcium channel, voltage-dependent, gamma subunit 1 |
| chr6_+_30851840 | 0.38 |
ENST00000511510.1 ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_156105878 | 0.38 |
ENST00000508500.1 |
LMNA |
lamin A/C |
| chr12_+_4714145 | 0.38 |
ENST00000545342.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr2_-_183903133 | 0.38 |
ENST00000361354.4 |
NCKAP1 |
NCK-associated protein 1 |
| chr1_-_242162375 | 0.37 |
ENST00000357246.3 |
MAP1LC3C |
microtubule-associated protein 1 light chain 3 gamma |
| chr2_-_225811747 | 0.37 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr1_+_39796810 | 0.35 |
ENST00000289893.4 |
MACF1 |
microtubule-actin crosslinking factor 1 |
| chr10_+_123872483 | 0.35 |
ENST00000369001.1 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr1_-_153363452 | 0.35 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr20_-_14318248 | 0.35 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr12_-_48152428 | 0.35 |
ENST00000449771.2 ENST00000395358.3 |
RAPGEF3 |
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr17_+_45286706 | 0.35 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr18_-_53068911 | 0.35 |
ENST00000537856.3 |
TCF4 |
transcription factor 4 |
| chr3_-_136471204 | 0.34 |
ENST00000480733.1 ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1 |
stromal antigen 1 |
| chr12_+_56114151 | 0.34 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr4_-_186733363 | 0.34 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr6_+_99282570 | 0.33 |
ENST00000328345.5 |
POU3F2 |
POU class 3 homeobox 2 |
| chr12_+_56114189 | 0.33 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr8_-_145016692 | 0.32 |
ENST00000357649.2 |
PLEC |
plectin |
| chrX_-_134049262 | 0.32 |
ENST00000370783.3 |
MOSPD1 |
motile sperm domain containing 1 |
| chr2_-_178257401 | 0.31 |
ENST00000464747.1 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr15_-_41694640 | 0.31 |
ENST00000558719.1 ENST00000260361.4 ENST00000560978.1 |
NDUFAF1 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
| chr1_-_226111929 | 0.30 |
ENST00000343818.6 ENST00000432920.2 |
PYCR2 RP4-559A3.7 |
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr17_-_1090599 | 0.30 |
ENST00000544583.2 |
ABR |
active BCR-related |
| chr3_-_178790057 | 0.29 |
ENST00000311417.2 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr6_+_25279651 | 0.28 |
ENST00000329474.6 |
LRRC16A |
leucine rich repeat containing 16A |
| chr3_+_183892635 | 0.28 |
ENST00000427072.1 ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
| chr5_-_150466692 | 0.28 |
ENST00000315050.7 ENST00000523338.1 ENST00000522100.1 |
TNIP1 |
TNFAIP3 interacting protein 1 |
| chr5_+_49962772 | 0.28 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr6_-_137113604 | 0.28 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr3_+_118905564 | 0.27 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chrX_-_13835461 | 0.27 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr5_-_171433819 | 0.26 |
ENST00000296933.6 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr2_-_152830441 | 0.26 |
ENST00000534999.1 ENST00000397327.2 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr19_-_4454081 | 0.26 |
ENST00000591919.1 |
UBXN6 |
UBX domain protein 6 |
| chr1_+_36023035 | 0.25 |
ENST00000373253.3 |
NCDN |
neurochondrin |
| chrX_+_150151752 | 0.25 |
ENST00000325307.7 |
HMGB3 |
high mobility group box 3 |
| chr8_+_98788003 | 0.25 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr19_+_41222998 | 0.24 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr17_-_3301704 | 0.23 |
ENST00000322608.2 |
OR1E1 |
olfactory receptor, family 1, subfamily E, member 1 |
| chr4_-_103748880 | 0.23 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr18_-_52989217 | 0.23 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr12_+_26348582 | 0.22 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr1_-_84464780 | 0.22 |
ENST00000260505.8 |
TTLL7 |
tubulin tyrosine ligase-like family, member 7 |
| chr8_+_9413410 | 0.22 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr7_+_18535346 | 0.22 |
ENST00000405010.3 ENST00000406451.4 ENST00000428307.2 |
HDAC9 |
histone deacetylase 9 |
| chr12_+_14518598 | 0.22 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr18_-_52989525 | 0.22 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chrX_-_41782683 | 0.22 |
ENST00000378163.1 ENST00000378154.1 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr9_-_128003606 | 0.21 |
ENST00000324460.6 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr11_+_8008867 | 0.21 |
ENST00000309828.4 ENST00000449102.2 |
EIF3F |
eukaryotic translation initiation factor 3, subunit F |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 2.4 | 9.8 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 1.6 | 6.4 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.8 | 3.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.7 | 2.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.7 | 7.4 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.5 | 3.8 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.5 | 2.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 1.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.3 | 1.8 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 1.7 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 0.5 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.2 | 0.7 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 2.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 0.7 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.2 | 1.1 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 | 0.3 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.2 | 1.4 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.4 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.4 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.1 | 1.7 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.1 | 1.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 1.6 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.1 | 1.0 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.8 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.1 | 1.0 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.3 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.7 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.1 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 0.3 | GO:1904753 | negative regulation of vascular associated smooth muscle cell migration(GO:1904753) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 1.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.4 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.5 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.2 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.7 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.1 | 4.9 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.1 | 1.1 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 1.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.6 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.6 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.3 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.1 | 0.7 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.7 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.2 | GO:0006853 | carnitine shuttle(GO:0006853) |
| 0.1 | 3.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.6 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 3.5 | GO:0033275 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.1 | 0.3 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.1 | 0.4 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.1 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 3.7 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 1.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1901899 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 1.4 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.6 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 2.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 1.1 | GO:1901685 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0072276 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.7 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 5.6 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0060513 | prostatic bud formation(GO:0060513) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.5 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.7 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 1.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.5 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 1.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.5 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.4 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 1.7 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.3 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.6 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 2.9 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.0 | 0.9 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.2 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.0 | GO:1902908 | regulation of melanosome transport(GO:1902908) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0071313 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.1 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 16.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 2.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.5 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 6.4 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.4 | 3.9 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 6.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 3.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 7.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 0.7 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 7.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.2 | 1.5 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 2.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 2.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 13.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 4.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.6 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 2.9 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.2 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.6 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.7 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.7 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 5.8 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 1.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 5.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 1.4 | GO:0031901 | early endosome membrane(GO:0031901) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.8 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.6 | 6.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.4 | 1.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.4 | 1.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 2.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.2 | 0.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.7 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.2 | 21.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.2 | 0.4 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 0.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 1.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 2.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 1.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 3.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.5 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.1 | 0.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.0 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.1 | 0.5 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.1 | 0.5 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 2.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.9 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 12.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 1.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.1 | 7.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 1.8 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.7 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 3.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.2 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 5.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0008301 | four-way junction DNA binding(GO:0000400) DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.5 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 1.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.0 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 8.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 9.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 12.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 7.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 2.6 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |