Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
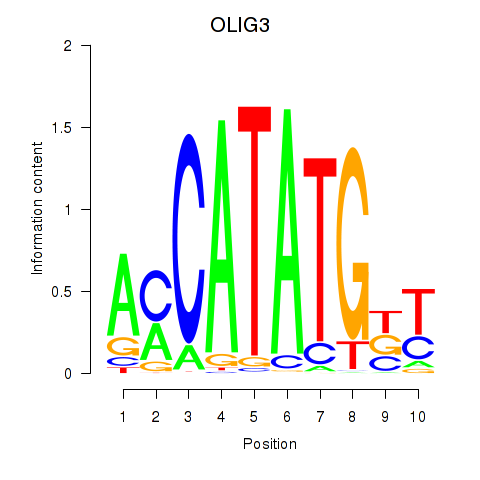
Results for OLIG3_NEUROD2_NEUROG2
Z-value: 1.13
Transcription factors associated with OLIG3_NEUROD2_NEUROG2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG3
|
ENSG00000177468.5 | OLIG3 |
|
NEUROD2
|
ENSG00000171532.4 | NEUROD2 |
|
NEUROG2
|
ENSG00000178403.3 | NEUROG2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROG2 | hg19_v2_chr4_-_113437328_113437337 | 0.02 | 9.4e-01 | Click! |
Activity profile of OLIG3_NEUROD2_NEUROG2 motif
Sorted Z-values of OLIG3_NEUROD2_NEUROG2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG3_NEUROD2_NEUROG2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_69597810 | 4.18 |
ENST00000483798.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr2_-_188378368 | 3.56 |
ENST00000392365.1 ENST00000435414.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr10_-_69597915 | 2.88 |
ENST00000225171.2 |
DNAJC12 |
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_+_5135981 | 2.39 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr12_-_91573132 | 2.33 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr2_+_11674213 | 2.29 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr6_+_3000057 | 2.06 |
ENST00000397717.2 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
| chr1_-_144994909 | 1.93 |
ENST00000369347.4 ENST00000369354.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr1_-_144995074 | 1.89 |
ENST00000534536.1 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr6_+_3000195 | 1.88 |
ENST00000338130.2 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
| chr6_+_3000218 | 1.77 |
ENST00000380441.1 ENST00000380455.4 ENST00000380454.4 |
NQO2 |
NAD(P)H dehydrogenase, quinone 2 |
| chr8_+_21823726 | 1.77 |
ENST00000433566.4 |
XPO7 |
exportin 7 |
| chr12_-_91573249 | 1.74 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr11_-_72070206 | 1.70 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr4_+_74301880 | 1.66 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr20_+_34802295 | 1.58 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr3_-_148939835 | 1.57 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr12_-_91573316 | 1.52 |
ENST00000393155.1 |
DCN |
decorin |
| chr11_+_59824060 | 1.41 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr1_-_144994840 | 1.41 |
ENST00000369351.3 ENST00000369349.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr2_-_166930131 | 1.40 |
ENST00000303395.4 ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A |
sodium channel, voltage-gated, type I, alpha subunit |
| chr16_-_55867146 | 1.37 |
ENST00000422046.2 |
CES1 |
carboxylesterase 1 |
| chr1_-_205091115 | 1.21 |
ENST00000264515.6 ENST00000367164.1 |
RBBP5 |
retinoblastoma binding protein 5 |
| chr1_-_144995002 | 1.17 |
ENST00000369356.4 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr6_+_135502501 | 1.15 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_-_114477962 | 1.11 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr12_+_57849048 | 1.09 |
ENST00000266646.2 |
INHBE |
inhibin, beta E |
| chr2_+_189839046 | 1.07 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr3_+_69928256 | 1.07 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr22_+_21133469 | 1.06 |
ENST00000406799.1 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr12_-_91546926 | 1.06 |
ENST00000550758.1 |
DCN |
decorin |
| chr3_-_178984759 | 1.04 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_-_114477787 | 1.02 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr19_-_49944806 | 0.99 |
ENST00000221485.3 |
SLC17A7 |
solute carrier family 17 (vesicular glutamate transporter), member 7 |
| chr5_+_157158205 | 0.93 |
ENST00000231198.7 |
THG1L |
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr9_-_130635741 | 0.92 |
ENST00000223836.10 |
AK1 |
adenylate kinase 1 |
| chr20_-_32031680 | 0.91 |
ENST00000217381.2 |
SNTA1 |
syntrophin, alpha 1 |
| chr21_-_34914394 | 0.91 |
ENST00000361093.5 ENST00000381815.4 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr15_-_80189380 | 0.90 |
ENST00000258874.3 |
MTHFS |
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr17_-_64216748 | 0.89 |
ENST00000585162.1 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr1_-_145076186 | 0.85 |
ENST00000369348.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr14_-_69864993 | 0.85 |
ENST00000555373.1 |
ERH |
enhancer of rudimentary homolog (Drosophila) |
| chr7_-_150652924 | 0.83 |
ENST00000330883.4 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr3_+_185300391 | 0.83 |
ENST00000545472.1 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr14_+_95047725 | 0.82 |
ENST00000554760.1 ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr4_+_69962185 | 0.80 |
ENST00000305231.7 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chrX_+_69509927 | 0.80 |
ENST00000374403.3 |
KIF4A |
kinesin family member 4A |
| chr15_+_65843130 | 0.79 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr17_-_7017559 | 0.79 |
ENST00000446679.2 |
ASGR2 |
asialoglycoprotein receptor 2 |
| chr4_+_69962212 | 0.79 |
ENST00000508661.1 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_-_160147925 | 0.78 |
ENST00000535561.1 |
SOD2 |
superoxide dismutase 2, mitochondrial |
| chr14_+_95047744 | 0.77 |
ENST00000553511.1 ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr6_+_37400974 | 0.77 |
ENST00000455891.1 ENST00000373451.4 |
CMTR1 |
cap methyltransferase 1 |
| chr19_+_19569270 | 0.76 |
ENST00000358713.3 |
GATAD2A |
GATA zinc finger domain containing 2A |
| chrX_-_122756660 | 0.74 |
ENST00000441692.1 |
THOC2 |
THO complex 2 |
| chr3_-_52486841 | 0.72 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr4_-_74088800 | 0.69 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr11_-_118213455 | 0.67 |
ENST00000300692.4 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr20_-_35274548 | 0.66 |
ENST00000262866.4 |
SLA2 |
Src-like-adaptor 2 |
| chr4_-_155533787 | 0.65 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr8_-_17752996 | 0.63 |
ENST00000381841.2 ENST00000427924.1 |
FGL1 |
fibrinogen-like 1 |
| chr14_-_21490958 | 0.63 |
ENST00000554104.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_21490590 | 0.62 |
ENST00000557633.1 |
NDRG2 |
NDRG family member 2 |
| chr4_-_89744457 | 0.62 |
ENST00000395002.2 |
FAM13A |
family with sequence similarity 13, member A |
| chr19_-_52227221 | 0.60 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr17_-_28618867 | 0.59 |
ENST00000394819.3 ENST00000577623.1 |
BLMH |
bleomycin hydrolase |
| chr15_+_63354769 | 0.59 |
ENST00000558910.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr4_+_144354644 | 0.58 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr9_-_75567962 | 0.57 |
ENST00000297785.3 ENST00000376939.1 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
| chr1_-_8000872 | 0.57 |
ENST00000377507.3 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
| chr14_-_21490653 | 0.55 |
ENST00000449431.2 |
NDRG2 |
NDRG family member 2 |
| chr1_-_23810664 | 0.54 |
ENST00000336689.3 ENST00000437606.2 |
ASAP3 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr1_+_145883868 | 0.54 |
ENST00000447947.2 |
GPR89C |
G protein-coupled receptor 89C |
| chr1_+_169764163 | 0.54 |
ENST00000413811.2 ENST00000359326.4 ENST00000456684.1 |
C1orf112 |
chromosome 1 open reading frame 112 |
| chr17_+_16318909 | 0.53 |
ENST00000577397.1 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
| chr4_-_141348999 | 0.53 |
ENST00000325617.5 |
CLGN |
calmegin |
| chr14_-_25479811 | 0.52 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr1_+_196621002 | 0.51 |
ENST00000367429.4 ENST00000439155.2 |
CFH |
complement factor H |
| chr11_+_7506713 | 0.50 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr17_+_41003166 | 0.50 |
ENST00000308423.2 |
AOC3 |
amine oxidase, copper containing 3 |
| chr10_-_5708515 | 0.49 |
ENST00000357700.6 |
ASB13 |
ankyrin repeat and SOCS box containing 13 |
| chr6_+_32146131 | 0.48 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr7_-_99573640 | 0.48 |
ENST00000411734.1 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr12_-_122296755 | 0.48 |
ENST00000289004.4 |
HPD |
4-hydroxyphenylpyruvate dioxygenase |
| chr17_+_16318850 | 0.48 |
ENST00000338560.7 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
| chr10_+_51565188 | 0.48 |
ENST00000430396.2 ENST00000374087.4 ENST00000414907.2 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr21_-_43786634 | 0.47 |
ENST00000291527.2 |
TFF1 |
trefoil factor 1 |
| chr14_-_21490417 | 0.47 |
ENST00000556366.1 |
NDRG2 |
NDRG family member 2 |
| chr8_+_21915368 | 0.47 |
ENST00000265800.5 ENST00000517418.1 |
DMTN |
dematin actin binding protein |
| chr10_+_51565108 | 0.47 |
ENST00000438493.1 ENST00000452682.1 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr12_+_100897130 | 0.46 |
ENST00000551379.1 ENST00000188403.7 ENST00000551184.1 |
NR1H4 |
nuclear receptor subfamily 1, group H, member 4 |
| chr1_+_146714291 | 0.46 |
ENST00000431239.1 ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
| chr5_+_53751445 | 0.46 |
ENST00000302005.1 |
HSPB3 |
heat shock 27kDa protein 3 |
| chr6_+_30585486 | 0.46 |
ENST00000259873.4 ENST00000506373.2 |
MRPS18B |
mitochondrial ribosomal protein S18B |
| chr7_-_92463210 | 0.46 |
ENST00000265734.4 |
CDK6 |
cyclin-dependent kinase 6 |
| chr6_+_30539153 | 0.45 |
ENST00000326195.8 ENST00000376545.3 ENST00000396515.4 ENST00000441867.1 ENST00000468958.1 |
ABCF1 |
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr1_+_54359854 | 0.44 |
ENST00000361921.3 ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1 |
deiodinase, iodothyronine, type I |
| chr4_-_174451370 | 0.44 |
ENST00000359562.4 |
HAND2 |
heart and neural crest derivatives expressed 2 |
| chr2_+_178257372 | 0.44 |
ENST00000264167.4 ENST00000409888.1 |
AGPS |
alkylglycerone phosphate synthase |
| chrX_-_154563889 | 0.44 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr5_-_137674000 | 0.43 |
ENST00000510119.1 ENST00000513970.1 |
CDC25C |
cell division cycle 25C |
| chr1_+_12524965 | 0.42 |
ENST00000471923.1 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr22_+_45072958 | 0.41 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr3_-_194072019 | 0.41 |
ENST00000429275.1 ENST00000323830.3 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
| chr6_-_28891709 | 0.41 |
ENST00000377194.3 ENST00000377199.3 |
TRIM27 |
tripartite motif containing 27 |
| chr1_+_46049706 | 0.41 |
ENST00000527470.1 ENST00000525515.1 ENST00000537798.1 ENST00000402363.3 ENST00000528238.1 ENST00000350030.3 ENST00000470768.1 ENST00000372052.4 ENST00000351223.3 |
NASP |
nuclear autoantigenic sperm protein (histone-binding) |
| chr19_+_18208603 | 0.40 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr7_-_97881429 | 0.40 |
ENST00000420697.1 ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1 |
tectonin beta-propeller repeat containing 1 |
| chr1_+_179050512 | 0.40 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr17_-_42345487 | 0.38 |
ENST00000262418.6 |
SLC4A1 |
solute carrier family 4 (anion exchanger), member 1 (Diego blood group) |
| chr22_+_45072925 | 0.38 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr15_+_62853562 | 0.38 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr3_-_33700933 | 0.37 |
ENST00000480013.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr12_+_93861282 | 0.37 |
ENST00000552217.1 ENST00000393128.4 ENST00000547098.1 |
MRPL42 |
mitochondrial ribosomal protein L42 |
| chr19_+_18496957 | 0.36 |
ENST00000252809.3 |
GDF15 |
growth differentiation factor 15 |
| chr10_-_75415825 | 0.36 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr11_-_118972575 | 0.36 |
ENST00000432443.2 |
DPAGT1 |
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr4_-_111563076 | 0.36 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr17_-_40428359 | 0.36 |
ENST00000293328.3 |
STAT5B |
signal transducer and activator of transcription 5B |
| chr1_-_155959853 | 0.35 |
ENST00000462460.2 ENST00000368316.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr12_+_93861264 | 0.35 |
ENST00000549982.1 ENST00000361630.2 |
MRPL42 |
mitochondrial ribosomal protein L42 |
| chr15_-_90358048 | 0.35 |
ENST00000300060.6 ENST00000560137.1 |
ANPEP |
alanyl (membrane) aminopeptidase |
| chr6_+_10528560 | 0.34 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr15_-_55700216 | 0.34 |
ENST00000569205.1 |
CCPG1 |
cell cycle progression 1 |
| chr7_-_99573677 | 0.33 |
ENST00000292401.4 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr5_-_75919217 | 0.33 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr9_+_134000948 | 0.33 |
ENST00000359428.5 ENST00000411637.2 ENST00000451030.1 |
NUP214 |
nucleoporin 214kDa |
| chr19_-_10445399 | 0.33 |
ENST00000592945.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr5_-_169725231 | 0.33 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr9_+_90341024 | 0.33 |
ENST00000340342.6 ENST00000342020.5 |
CTSL |
cathepsin L |
| chr2_+_105953972 | 0.32 |
ENST00000410049.1 |
C2orf49 |
chromosome 2 open reading frame 49 |
| chr1_-_71513471 | 0.32 |
ENST00000370931.3 ENST00000356595.4 ENST00000306666.5 ENST00000370932.2 ENST00000351052.5 ENST00000414819.1 ENST00000370924.4 |
PTGER3 |
prostaglandin E receptor 3 (subtype EP3) |
| chr15_-_55700522 | 0.32 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr15_-_49447835 | 0.32 |
ENST00000388901.5 ENST00000299259.6 |
COPS2 |
COP9 signalosome subunit 2 |
| chr17_+_56769924 | 0.32 |
ENST00000461271.1 ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C |
RAD51 paralog C |
| chr3_+_157828152 | 0.32 |
ENST00000476899.1 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
| chr19_-_15529790 | 0.32 |
ENST00000596195.1 ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L |
A kinase (PRKA) anchor protein 8-like |
| chr5_-_75919253 | 0.32 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr2_-_152382500 | 0.31 |
ENST00000434685.1 |
NEB |
nebulin |
| chr6_-_32145861 | 0.31 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr4_-_70080449 | 0.31 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr10_-_97416400 | 0.31 |
ENST00000371224.2 ENST00000371221.3 |
ALDH18A1 |
aldehyde dehydrogenase 18 family, member A1 |
| chr19_-_47291843 | 0.31 |
ENST00000542575.2 |
SLC1A5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr15_+_74466012 | 0.31 |
ENST00000249842.3 |
ISLR |
immunoglobulin superfamily containing leucine-rich repeat |
| chrX_+_118108601 | 0.30 |
ENST00000371628.3 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chr7_+_90893783 | 0.30 |
ENST00000287934.2 |
FZD1 |
frizzled family receptor 1 |
| chr22_+_24999114 | 0.30 |
ENST00000412658.1 ENST00000445029.1 ENST00000419133.1 ENST00000400382.1 ENST00000438643.2 ENST00000452551.1 ENST00000400383.1 ENST00000412898.1 ENST00000400380.1 ENST00000455483.1 ENST00000430289.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr7_-_140482926 | 0.30 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr1_+_156182773 | 0.30 |
ENST00000490491.1 ENST00000368276.4 ENST00000320139.5 ENST00000368279.3 ENST00000368273.4 ENST00000368277.3 ENST00000567140.1 ENST00000565805.1 |
PMF1-BGLAP PMF1 |
PMF1-BGLAP readthrough polyamine-modulated factor 1 |
| chrX_+_118108571 | 0.29 |
ENST00000304778.7 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chr1_-_111970353 | 0.29 |
ENST00000369732.3 |
OVGP1 |
oviductal glycoprotein 1, 120kDa |
| chr1_+_235492300 | 0.29 |
ENST00000476121.1 ENST00000497327.1 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chr12_+_113376249 | 0.29 |
ENST00000551007.1 ENST00000548514.1 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr4_-_89744365 | 0.29 |
ENST00000513837.1 ENST00000503556.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr1_+_15943995 | 0.29 |
ENST00000480945.1 |
DDI2 |
DNA-damage inducible 1 homolog 2 (S. cerevisiae) |
| chr17_+_72426891 | 0.28 |
ENST00000392627.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_89990431 | 0.28 |
ENST00000330947.2 ENST00000358200.4 |
LRRC8B |
leucine rich repeat containing 8 family, member B |
| chr1_+_57320437 | 0.28 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr4_-_186733363 | 0.28 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr11_-_11374904 | 0.28 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
| chr12_-_131323754 | 0.28 |
ENST00000261653.6 |
STX2 |
syntaxin 2 |
| chr17_+_80416482 | 0.28 |
ENST00000309794.11 ENST00000345415.7 ENST00000457415.3 ENST00000584411.1 ENST00000412079.2 ENST00000577432.1 |
NARF |
nuclear prelamin A recognition factor |
| chrX_+_114423963 | 0.28 |
ENST00000424776.3 |
RBMXL3 |
RNA binding motif protein, X-linked-like 3 |
| chr7_+_105172532 | 0.27 |
ENST00000257700.2 |
RINT1 |
RAD50 interactor 1 |
| chr9_-_127177703 | 0.27 |
ENST00000259457.3 ENST00000536392.1 ENST00000441097.1 |
PSMB7 |
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr1_-_44818599 | 0.27 |
ENST00000537474.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr2_+_241807870 | 0.27 |
ENST00000307503.3 |
AGXT |
alanine-glyoxylate aminotransferase |
| chr5_+_43602750 | 0.27 |
ENST00000505678.2 ENST00000512422.1 ENST00000264663.5 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chrX_+_120181457 | 0.27 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr5_+_135496675 | 0.27 |
ENST00000507637.1 |
SMAD5 |
SMAD family member 5 |
| chr4_-_89744314 | 0.26 |
ENST00000508369.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr10_+_102747783 | 0.26 |
ENST00000311916.2 ENST00000370228.1 |
C10orf2 |
chromosome 10 open reading frame 2 |
| chr14_-_23904861 | 0.26 |
ENST00000355349.3 |
MYH7 |
myosin, heavy chain 7, cardiac muscle, beta |
| chr1_-_220219775 | 0.26 |
ENST00000609181.1 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr3_-_170744498 | 0.26 |
ENST00000382808.4 ENST00000314251.3 |
SLC2A2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr2_-_163099885 | 0.26 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr15_-_72668185 | 0.26 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr2_-_40006357 | 0.25 |
ENST00000505747.1 |
THUMPD2 |
THUMP domain containing 2 |
| chr3_+_139063372 | 0.25 |
ENST00000478464.1 |
MRPS22 |
mitochondrial ribosomal protein S22 |
| chr4_+_71063641 | 0.25 |
ENST00000514097.1 |
ODAM |
odontogenic, ameloblast asssociated |
| chr4_+_71600144 | 0.25 |
ENST00000502653.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr9_-_123239632 | 0.25 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr16_-_20364122 | 0.25 |
ENST00000396138.4 ENST00000577168.1 |
UMOD |
uromodulin |
| chr17_+_66511540 | 0.25 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr16_-_20364030 | 0.25 |
ENST00000396134.2 ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD |
uromodulin |
| chr2_-_40006289 | 0.24 |
ENST00000260619.6 ENST00000454352.2 |
THUMPD2 |
THUMP domain containing 2 |
| chr17_+_34842473 | 0.24 |
ENST00000490126.2 ENST00000225410.4 |
ZNHIT3 |
zinc finger, HIT-type containing 3 |
| chr11_-_88796803 | 0.23 |
ENST00000418177.2 ENST00000455756.2 |
GRM5 |
glutamate receptor, metabotropic 5 |
| chr12_+_6898638 | 0.23 |
ENST00000011653.4 |
CD4 |
CD4 molecule |
| chr3_-_33700589 | 0.23 |
ENST00000461133.3 ENST00000496954.2 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr10_-_5446786 | 0.23 |
ENST00000479328.1 ENST00000380419.3 |
TUBAL3 |
tubulin, alpha-like 3 |
| chr1_+_11994715 | 0.23 |
ENST00000449038.1 ENST00000376369.3 ENST00000429000.2 ENST00000196061.4 |
PLOD1 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 |
| chr17_-_61850894 | 0.23 |
ENST00000403162.3 ENST00000582252.1 ENST00000225726.5 |
CCDC47 |
coiled-coil domain containing 47 |
| chr20_-_16554078 | 0.23 |
ENST00000354981.2 ENST00000355755.3 ENST00000378003.2 ENST00000408042.1 |
KIF16B |
kinesin family member 16B |
| chr6_+_69942298 | 0.23 |
ENST00000238918.8 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr13_+_50656307 | 0.23 |
ENST00000378180.4 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr1_+_228337553 | 0.23 |
ENST00000366714.2 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
| chr17_-_15168624 | 0.23 |
ENST00000312280.3 ENST00000494511.1 ENST00000580584.1 |
PMP22 |
peripheral myelin protein 22 |
| chr1_+_159272111 | 0.23 |
ENST00000368114.1 |
FCER1A |
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr2_+_166428839 | 0.22 |
ENST00000342316.4 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0036030 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.5 | 6.6 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.2 | 0.8 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 1.0 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.1 | 0.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 1.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.7 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.3 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.1 | 1.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.3 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.4 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.1 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.1 | 0.3 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.9 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.7 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.6 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.4 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.9 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 4.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 8.3 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 4.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.2 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.7 | GO:1904408 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.6 | 2.6 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.4 | 1.8 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.3 | 0.9 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.3 | 1.4 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.2 | 1.0 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.2 | 2.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 0.8 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.6 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 0.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 1.6 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 0.5 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.2 | 0.5 | GO:1902122 | chenodeoxycholic acid binding(GO:1902122) |
| 0.2 | 0.8 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.2 | 0.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.1 | 0.8 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.7 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.6 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 1.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.8 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 1.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 6.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.8 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.3 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.8 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.3 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.2 | GO:0004961 | thromboxane receptor activity(GO:0004960) thromboxane A2 receptor activity(GO:0004961) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.1 | 0.2 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.6 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.8 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.5 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.7 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 1.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.2 | GO:0033989 | 3alpha,7alpha,12alpha-trihydroxy-5beta-cholest-24-enoyl-CoA hydratase activity(GO:0033989) 17-beta-hydroxysteroid dehydrogenase (NAD+) activity(GO:0044594) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.2 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.2 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.4 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.3 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.3 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 1.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 1.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.6 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 2.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.5 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.8 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.9 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 1.1 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.8 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 1.0 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.6 | 6.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 3.6 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.4 | 1.1 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.3 | 1.0 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.3 | 1.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.3 | 0.8 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 0.9 | GO:0015942 | folic acid-containing compound catabolic process(GO:0009397) formate metabolic process(GO:0015942) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 5.7 | GO:1904707 | positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.2 | 0.6 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.2 | 0.7 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 0.7 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.8 | GO:0003069 | age-dependent response to oxidative stress(GO:0001306) age-dependent response to reactive oxygen species(GO:0001315) regulation of systemic arterial blood pressure by acetylcholine(GO:0003068) vasodilation by acetylcholine involved in regulation of systemic arterial blood pressure(GO:0003069) regulation of systemic arterial blood pressure by neurotransmitter(GO:0003070) age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.5 | GO:1903314 | nitrogen catabolite regulation of transcription from RNA polymerase II promoter(GO:0001079) nitrogen catabolite activation of transcription from RNA polymerase II promoter(GO:0001080) regulation of urea metabolic process(GO:0034255) intracellular bile acid receptor signaling pathway(GO:0038185) interleukin-17 secretion(GO:0072615) nitrogen catabolite regulation of transcription(GO:0090293) nitrogen catabolite activation of transcription(GO:0090294) regulation of nitrogen cycle metabolic process(GO:1903314) positive regulation of glutamate metabolic process(GO:2000213) regulation of ammonia assimilation cycle(GO:2001248) positive regulation of ammonia assimilation cycle(GO:2001250) |
| 0.2 | 0.8 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.4 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.3 | GO:0009093 | cysteine catabolic process(GO:0009093) L-cysteine catabolic process(GO:0019448) L-cysteine metabolic process(GO:0046439) |
| 0.1 | 1.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 | 0.5 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.1 | 0.4 | GO:0060460 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 0.5 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.1 | 1.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.1 | 0.9 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.1 | 1.7 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.7 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.5 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.1 | 1.6 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.6 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.1 | 0.3 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.3 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 0.6 | GO:2001183 | negative regulation of interleukin-10 secretion(GO:2001180) negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.3 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.4 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.9 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.4 | GO:0003253 | cardiac right ventricle formation(GO:0003219) cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.3 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.4 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.5 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.5 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.3 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.1 | 0.5 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 0.6 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.1 | 0.3 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.1 | 0.6 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.9 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 0.5 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.3 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.1 | 0.2 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 1.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 | 0.2 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.1 | 1.6 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.4 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.2 | GO:0038193 | thromboxane A2 signaling pathway(GO:0038193) |
| 0.1 | 0.5 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 1.0 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.9 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.9 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.2 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.3 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.2 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.1 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.7 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.9 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.0 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.2 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.7 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.8 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.1 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 1.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.8 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.0 | 0.1 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 1.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.0 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 1.2 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.1 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 1.1 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 1.8 | GO:0070126 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.4 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.6 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.2 | GO:0097039 | CD40 signaling pathway(GO:0023035) protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 1.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.3 | GO:0002223 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 1.1 | GO:0090280 | positive regulation of calcium ion import(GO:0090280) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.3 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.3 | GO:0071888 | macrophage apoptotic process(GO:0071888) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.5 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0072757 | cellular response to camptothecin(GO:0072757) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.9 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.4 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.1 | GO:1903895 | negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.0 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.4 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 0.8 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0051354 | negative regulation of oxidoreductase activity(GO:0051354) |
| 0.0 | 0.5 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0098838 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.0 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.0 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.0 | GO:1904529 | regulation of actin filament binding(GO:1904529) negative regulation of actin filament binding(GO:1904530) regulation of actin binding(GO:1904616) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.3 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 1.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.3 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.1 | GO:0008582 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.0 | 0.1 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 0.1 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |