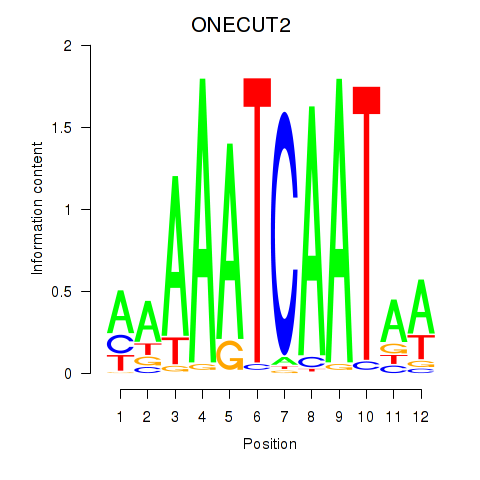
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for ONECUT2_ONECUT3
Z-value: 0.48
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | ONECUT2 |
|
ONECUT3
|
ENSG00000205922.4 | ONECUT3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT3 | hg19_v2_chr19_+_1752372_1752372 | 0.28 | 3.0e-01 | Click! |
| ONECUT2 | hg19_v2_chr18_+_55102917_55102985 | 0.17 | 5.2e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_4385230 | 1.46 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr1_+_198608146 | 0.77 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr11_-_107729887 | 0.58 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr7_-_23510086 | 0.54 |
ENST00000258729.3 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
| chr7_-_100239132 | 0.50 |
ENST00000223051.3 ENST00000431692.1 |
TFR2 |
transferrin receptor 2 |
| chr9_-_27529726 | 0.50 |
ENST00000262244.5 |
MOB3B |
MOB kinase activator 3B |
| chr6_+_56954867 | 0.33 |
ENST00000370708.4 ENST00000370702.1 |
ZNF451 |
zinc finger protein 451 |
| chr6_-_41715128 | 0.32 |
ENST00000356667.4 ENST00000373025.3 ENST00000425343.2 |
PGC |
progastricsin (pepsinogen C) |
| chr3_-_24207039 | 0.32 |
ENST00000280696.5 |
THRB |
thyroid hormone receptor, beta |
| chr9_-_138853156 | 0.30 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chr14_-_23426337 | 0.28 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr14_-_23426270 | 0.28 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr21_-_15918618 | 0.27 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr14_-_23426322 | 0.27 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr10_-_98031310 | 0.27 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr2_+_58655461 | 0.26 |
ENST00000429095.1 ENST00000429664.1 ENST00000452840.1 |
AC007092.1 |
long intergenic non-protein coding RNA 1122 |
| chr10_-_98031265 | 0.26 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr6_+_56954808 | 0.24 |
ENST00000510483.1 ENST00000370706.4 ENST00000357489.3 |
ZNF451 |
zinc finger protein 451 |
| chr10_-_62332357 | 0.24 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr15_-_37392703 | 0.24 |
ENST00000382766.2 ENST00000444725.1 |
MEIS2 |
Meis homeobox 2 |
| chr20_-_62587735 | 0.24 |
ENST00000354216.6 ENST00000369892.3 ENST00000358711.3 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
| chrX_-_133792480 | 0.23 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr3_+_121774202 | 0.23 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr12_+_59989918 | 0.23 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_-_192016316 | 0.23 |
ENST00000358470.4 ENST00000432798.1 ENST00000450994.1 |
STAT4 |
signal transducer and activator of transcription 4 |
| chr20_+_36405665 | 0.23 |
ENST00000373469.1 |
CTNNBL1 |
catenin, beta like 1 |
| chr17_-_41623691 | 0.22 |
ENST00000545954.1 |
ETV4 |
ets variant 4 |
| chr5_-_115910630 | 0.22 |
ENST00000343348.6 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr11_-_35440796 | 0.22 |
ENST00000278379.3 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_39348137 | 0.21 |
ENST00000426016.1 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr18_+_42260861 | 0.21 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr17_-_64225508 | 0.20 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr14_+_37126765 | 0.20 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr15_-_37392086 | 0.19 |
ENST00000561208.1 |
MEIS2 |
Meis homeobox 2 |
| chr10_-_121296045 | 0.19 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr1_-_116383322 | 0.19 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr20_-_22566089 | 0.19 |
ENST00000377115.4 |
FOXA2 |
forkhead box A2 |
| chr17_-_41623716 | 0.19 |
ENST00000319349.5 |
ETV4 |
ets variant 4 |
| chr14_+_39736582 | 0.18 |
ENST00000556148.1 ENST00000348007.3 |
CTAGE5 |
CTAGE family, member 5 |
| chr16_+_72088376 | 0.18 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chr1_-_116383738 | 0.18 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr7_-_104909435 | 0.16 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr21_+_39628655 | 0.15 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr18_+_11752040 | 0.15 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr15_+_93443419 | 0.15 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr1_+_207943667 | 0.15 |
ENST00000462968.2 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr10_+_133753533 | 0.15 |
ENST00000422256.2 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
| chr1_-_32384693 | 0.15 |
ENST00000602683.1 ENST00000470404.1 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
| chr21_+_39628852 | 0.15 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr19_+_37998031 | 0.15 |
ENST00000586138.1 ENST00000588578.1 ENST00000587986.1 |
ZNF793 |
zinc finger protein 793 |
| chr3_-_141747439 | 0.14 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_894620 | 0.14 |
ENST00000327044.6 |
NOC2L |
nucleolar complex associated 2 homolog (S. cerevisiae) |
| chr2_-_208030647 | 0.13 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr9_-_70490107 | 0.12 |
ENST00000377395.4 ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5 |
COBW domain containing 5 |
| chr19_+_7598890 | 0.12 |
ENST00000221249.6 ENST00000601668.1 ENST00000601001.1 |
PNPLA6 |
patatin-like phospholipase domain containing 6 |
| chr9_+_70856397 | 0.11 |
ENST00000360171.6 |
CBWD3 |
COBW domain containing 3 |
| chr1_+_207262540 | 0.11 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr3_+_171561127 | 0.11 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chr10_+_95517616 | 0.11 |
ENST00000371418.4 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr6_+_37400974 | 0.11 |
ENST00000455891.1 ENST00000373451.4 |
CMTR1 |
cap methyltransferase 1 |
| chr18_+_29671812 | 0.11 |
ENST00000261593.3 ENST00000578914.1 |
RNF138 |
ring finger protein 138, E3 ubiquitin protein ligase |
| chr7_+_80231466 | 0.10 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr12_+_41831485 | 0.10 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr11_-_35440579 | 0.10 |
ENST00000606205.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_-_141747459 | 0.10 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr16_-_8962544 | 0.10 |
ENST00000570125.1 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chr17_+_7461849 | 0.10 |
ENST00000338784.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_117297007 | 0.10 |
ENST00000369478.3 ENST00000369477.1 |
CD2 |
CD2 molecule |
| chr12_+_59989791 | 0.10 |
ENST00000552432.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_-_50201327 | 0.10 |
ENST00000412315.1 |
NRXN1 |
neurexin 1 |
| chr10_+_95517566 | 0.10 |
ENST00000542308.1 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr5_+_65440032 | 0.09 |
ENST00000334121.6 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
| chr1_+_207262578 | 0.09 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr11_+_8040739 | 0.09 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
| chr2_+_157330081 | 0.09 |
ENST00000409674.1 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr10_+_70980051 | 0.09 |
ENST00000354624.5 ENST00000395086.2 |
HKDC1 |
hexokinase domain containing 1 |
| chr12_+_9144626 | 0.09 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr12_+_32832134 | 0.09 |
ENST00000452533.2 |
DNM1L |
dynamin 1-like |
| chr1_-_151431647 | 0.09 |
ENST00000368863.2 ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ |
pogo transposable element with ZNF domain |
| chr1_+_207262881 | 0.09 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr2_+_166095898 | 0.08 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr7_+_129007964 | 0.08 |
ENST00000460109.1 ENST00000474594.1 ENST00000446212.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr7_-_143059780 | 0.08 |
ENST00000409578.1 ENST00000409346.1 |
FAM131B |
family with sequence similarity 131, member B |
| chr12_+_86268065 | 0.08 |
ENST00000551529.1 ENST00000256010.6 |
NTS |
neurotensin |
| chr12_+_32832203 | 0.08 |
ENST00000553257.1 ENST00000549701.1 ENST00000358214.5 ENST00000266481.6 ENST00000551476.1 ENST00000550154.1 ENST00000547312.1 ENST00000414834.2 ENST00000381000.4 ENST00000548750.1 |
DNM1L |
dynamin 1-like |
| chr3_-_170744498 | 0.08 |
ENST00000382808.4 ENST00000314251.3 |
SLC2A2 |
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr12_+_81471816 | 0.08 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr16_-_8962200 | 0.08 |
ENST00000562843.1 ENST00000561530.1 ENST00000396593.2 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chrY_+_27768264 | 0.08 |
ENST00000361963.2 ENST00000306609.4 |
CDY1 |
chromodomain protein, Y-linked, 1 |
| chr7_-_143059845 | 0.08 |
ENST00000443739.2 |
FAM131B |
family with sequence similarity 131, member B |
| chr8_+_31497271 | 0.08 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chrX_+_123480194 | 0.08 |
ENST00000371139.4 |
SH2D1A |
SH2 domain containing 1A |
| chr1_+_207262627 | 0.08 |
ENST00000391923.1 |
C4BPB |
complement component 4 binding protein, beta |
| chr7_+_39125365 | 0.08 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr2_-_113012592 | 0.07 |
ENST00000272570.5 ENST00000409573.2 |
ZC3H8 |
zinc finger CCCH-type containing 8 |
| chrY_-_26194116 | 0.07 |
ENST00000306882.4 ENST00000382407.1 |
CDY1B |
chromodomain protein, Y-linked, 1B |
| chr19_+_3880581 | 0.07 |
ENST00000450849.2 ENST00000301260.6 ENST00000398448.3 |
ATCAY |
ataxia, cerebellar, Cayman type |
| chr3_+_39424828 | 0.07 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chr8_-_86290333 | 0.07 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr1_-_151431909 | 0.07 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr1_-_217262969 | 0.07 |
ENST00000361525.3 |
ESRRG |
estrogen-related receptor gamma |
| chr3_-_155394152 | 0.07 |
ENST00000494598.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr19_+_45596398 | 0.07 |
ENST00000544069.2 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
| chr5_+_140800638 | 0.06 |
ENST00000398587.2 ENST00000518882.1 |
PCDHGA11 |
protocadherin gamma subfamily A, 11 |
| chr2_+_168725458 | 0.06 |
ENST00000392690.3 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr17_+_7461781 | 0.06 |
ENST00000349228.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chrX_-_16887963 | 0.06 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr7_+_142829162 | 0.06 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr3_-_155394099 | 0.06 |
ENST00000414191.1 |
PLCH1 |
phospholipase C, eta 1 |
| chr19_+_7599128 | 0.06 |
ENST00000545201.2 |
PNPLA6 |
patatin-like phospholipase domain containing 6 |
| chr3_+_134514093 | 0.06 |
ENST00000398015.3 |
EPHB1 |
EPH receptor B1 |
| chr2_-_206950781 | 0.06 |
ENST00000403263.1 |
INO80D |
INO80 complex subunit D |
| chr11_+_6947647 | 0.06 |
ENST00000278319.5 |
ZNF215 |
zinc finger protein 215 |
| chr7_+_129906660 | 0.05 |
ENST00000222481.4 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
| chr1_-_115259337 | 0.05 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr1_-_182921119 | 0.05 |
ENST00000423786.1 |
SHCBP1L |
SHC SH2-domain binding protein 1-like |
| chr11_-_102651343 | 0.05 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr17_-_46691990 | 0.05 |
ENST00000576562.1 |
HOXB8 |
homeobox B8 |
| chr7_-_14026063 | 0.05 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr13_+_97928395 | 0.05 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr16_-_66952742 | 0.05 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr7_-_112727774 | 0.05 |
ENST00000297146.3 ENST00000501255.2 |
GPR85 |
G protein-coupled receptor 85 |
| chr17_-_39623681 | 0.05 |
ENST00000225899.3 |
KRT32 |
keratin 32 |
| chr2_+_170440902 | 0.05 |
ENST00000448752.2 ENST00000418888.1 ENST00000414307.1 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
| chr3_+_130650738 | 0.05 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr6_-_79787902 | 0.05 |
ENST00000275034.4 |
PHIP |
pleckstrin homology domain interacting protein |
| chr19_-_4535233 | 0.04 |
ENST00000381848.3 ENST00000588887.1 ENST00000586133.1 |
PLIN5 |
perilipin 5 |
| chr12_+_100594557 | 0.04 |
ENST00000546902.1 ENST00000552376.1 ENST00000551617.1 |
ACTR6 |
ARP6 actin-related protein 6 homolog (yeast) |
| chr2_+_170440844 | 0.04 |
ENST00000260970.3 ENST00000433207.1 ENST00000409714.3 ENST00000462903.1 |
PPIG |
peptidylprolyl isomerase G (cyclophilin G) |
| chr1_-_217262933 | 0.04 |
ENST00000359162.2 |
ESRRG |
estrogen-related receptor gamma |
| chr1_+_159557607 | 0.04 |
ENST00000255040.2 |
APCS |
amyloid P component, serum |
| chr4_+_187187098 | 0.04 |
ENST00000403665.2 ENST00000264692.4 |
F11 |
coagulation factor XI |
| chr17_+_7461613 | 0.04 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr16_-_66952779 | 0.04 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr9_+_33629119 | 0.03 |
ENST00000331828.4 |
TRBV21OR9-2 |
T cell receptor beta variable 21/OR9-2 (pseudogene) |
| chr10_+_95517660 | 0.03 |
ENST00000371413.3 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr5_-_135701164 | 0.03 |
ENST00000355180.3 ENST00000426057.2 ENST00000513104.1 |
TRPC7 |
transient receptor potential cation channel, subfamily C, member 7 |
| chr16_-_20702578 | 0.03 |
ENST00000307493.4 ENST00000219151.4 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chrX_+_101478829 | 0.03 |
ENST00000372763.1 ENST00000372758.1 |
NXF2 |
nuclear RNA export factor 2 |
| chrY_-_19992098 | 0.03 |
ENST00000544303.1 ENST00000382867.3 |
CDY2B |
chromodomain protein, Y-linked, 2B |
| chr3_-_194119083 | 0.03 |
ENST00000401815.1 |
GP5 |
glycoprotein V (platelet) |
| chrY_+_20137667 | 0.03 |
ENST00000250838.4 ENST00000426790.1 |
CDY2A |
chromodomain protein, Y-linked, 2A |
| chr17_-_79876010 | 0.03 |
ENST00000328666.6 |
SIRT7 |
sirtuin 7 |
| chr2_-_220174166 | 0.03 |
ENST00000409251.3 ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr14_+_37131058 | 0.03 |
ENST00000361487.6 |
PAX9 |
paired box 9 |
| chr5_+_162887556 | 0.03 |
ENST00000393915.4 ENST00000432118.2 ENST00000358715.3 |
HMMR |
hyaluronan-mediated motility receptor (RHAMM) |
| chr19_+_13135386 | 0.02 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr3_-_187388173 | 0.02 |
ENST00000287641.3 |
SST |
somatostatin |
| chr17_-_46692287 | 0.02 |
ENST00000239144.4 |
HOXB8 |
homeobox B8 |
| chr8_+_77593448 | 0.02 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr17_-_67264947 | 0.02 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr12_-_4488872 | 0.02 |
ENST00000237837.1 |
FGF23 |
fibroblast growth factor 23 |
| chr6_+_29068386 | 0.02 |
ENST00000377171.3 |
OR2J1 |
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr6_-_151773232 | 0.02 |
ENST00000444024.1 ENST00000367303.4 |
RMND1 |
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr5_-_9630463 | 0.02 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr3_+_52811596 | 0.02 |
ENST00000542827.1 ENST00000273283.2 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr10_+_114710516 | 0.02 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrX_-_33229636 | 0.02 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr19_+_45596218 | 0.02 |
ENST00000421905.1 ENST00000221462.4 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
| chr20_+_43849941 | 0.02 |
ENST00000372769.3 |
SEMG2 |
semenogelin II |
| chr2_+_171034646 | 0.02 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr16_-_4817129 | 0.01 |
ENST00000545009.1 ENST00000219478.6 |
ZNF500 |
zinc finger protein 500 |
| chr4_-_103747011 | 0.01 |
ENST00000350435.7 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chrX_+_70586082 | 0.01 |
ENST00000373790.4 ENST00000449580.1 ENST00000423759.1 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
| chrX_-_19988382 | 0.01 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr10_-_73975657 | 0.01 |
ENST00000394919.1 ENST00000526751.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chr1_-_159684371 | 0.01 |
ENST00000255030.5 ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP |
C-reactive protein, pentraxin-related |
| chr7_+_99425633 | 0.01 |
ENST00000354829.2 ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43 |
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr11_+_63137251 | 0.01 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr5_-_160279207 | 0.01 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr4_+_187148556 | 0.01 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr9_-_95244781 | 0.01 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chrX_+_46771848 | 0.00 |
ENST00000218343.4 |
PHF16 |
jade family PHD finger 3 |
| chr12_+_48866448 | 0.00 |
ENST00000266594.1 |
ANP32D |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member D |
| chr3_-_11685345 | 0.00 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr4_-_100212132 | 0.00 |
ENST00000209668.2 |
ADH1A |
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr14_+_23012122 | 0.00 |
ENST00000390534.1 |
TRAJ3 |
T cell receptor alpha joining 3 |
| chr6_+_27925019 | 0.00 |
ENST00000244623.1 |
OR2B6 |
olfactory receptor, family 2, subfamily B, member 6 |
| chr19_+_9361606 | 0.00 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr10_+_71562180 | 0.00 |
ENST00000517713.1 ENST00000522165.1 ENST00000520133.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr9_+_77230499 | 0.00 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr4_-_102267953 | 0.00 |
ENST00000523694.2 ENST00000507176.1 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr4_-_103746924 | 0.00 |
ENST00000505207.1 ENST00000502404.1 ENST00000507845.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.1 | 0.3 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.5 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.2 | GO:0032641 | negative regulation of tolerance induction(GO:0002644) lymphotoxin A production(GO:0032641) interleukin-4 biosynthetic process(GO:0042097) lymphotoxin A biosynthetic process(GO:0042109) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.1 | 0.6 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.1 | 1.5 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.2 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 0.2 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.2 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.1 | 0.2 | GO:0090149 | synaptic vesicle recycling via endosome(GO:0036466) mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.3 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.2 | GO:1904693 | midbrain morphogenesis(GO:1904693) |
| 0.0 | 0.4 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.3 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.2 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.4 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.0 | 0.2 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.1 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.8 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0035473 | lipase binding(GO:0035473) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |