Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
Z-value: 0.82
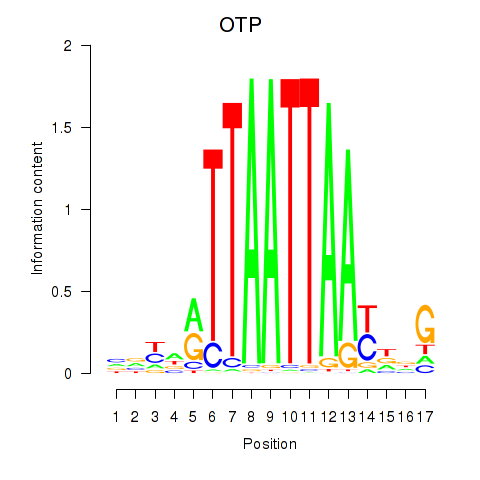
Transcription factors associated with OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTP
|
ENSG00000171540.6 | OTP |
|
PHOX2B
|
ENSG00000109132.5 | PHOX2B |
|
LHX1
|
ENSG00000132130.7 | LHX1 |
|
LMX1A
|
ENSG00000162761.10 | LMX1A |
|
LHX5
|
ENSG00000089116.3 | LHX5 |
|
HOXC4
|
ENSG00000198353.6 | HOXC4 |
|
HOXC4
|
ENSG00000273266.1 | HOXC4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC4 | hg19_v2_chr12_+_54447637_54447661 | -0.53 | 3.6e-02 | Click! |
| LHX1 | hg19_v2_chr17_+_35294075_35294102 | -0.52 | 3.9e-02 | Click! |
| PHOX2B | hg19_v2_chr4_-_41750922_41750987 | -0.21 | 4.2e-01 | Click! |
| LHX5 | hg19_v2_chr12_-_113909877_113909877 | -0.03 | 9.2e-01 | Click! |
Activity profile of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
Sorted Z-values of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OTP_PHOX2B_LHX1_LMX1A_LHX5_HOXC4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_1674982 | 5.82 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr16_-_28634874 | 2.91 |
ENST00000395609.1 ENST00000350842.4 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr19_+_50016610 | 2.83 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr19_+_50016411 | 2.82 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr5_+_95066823 | 2.28 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr3_+_157154578 | 2.06 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr7_+_134528635 | 1.71 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr11_-_63376013 | 1.56 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr12_+_26348246 | 1.55 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr10_+_94451574 | 1.52 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr7_+_138145076 | 1.28 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr17_-_64225508 | 1.28 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr16_-_30122717 | 1.21 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr20_-_33735070 | 1.15 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr7_-_87342564 | 1.10 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr12_+_26348429 | 1.09 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr1_-_92371839 | 1.00 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr15_+_58702742 | 1.00 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr11_+_114168085 | 0.99 |
ENST00000541754.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr3_-_149293990 | 0.92 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr7_+_102553430 | 0.91 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr12_+_26348582 | 0.85 |
ENST00000535504.1 |
SSPN |
sarcospan |
| chr1_+_117544366 | 0.72 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr10_-_75415825 | 0.72 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr1_-_57045228 | 0.69 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr7_-_143599207 | 0.69 |
ENST00000355951.2 ENST00000479870.1 ENST00000478172.1 |
FAM115A |
family with sequence similarity 115, member A |
| chr4_+_155484155 | 0.66 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr2_+_152214098 | 0.60 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr7_-_107642348 | 0.59 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chr19_-_51920952 | 0.58 |
ENST00000356298.5 ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
| chr3_-_114477962 | 0.52 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr2_-_190927447 | 0.52 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr3_-_114477787 | 0.50 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr12_+_64798095 | 0.50 |
ENST00000332707.5 |
XPOT |
exportin, tRNA |
| chr3_-_185538849 | 0.50 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr11_-_327537 | 0.48 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr4_-_185275104 | 0.47 |
ENST00000317596.3 |
RP11-290F5.2 |
RP11-290F5.2 |
| chr3_-_99569821 | 0.46 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr4_+_155484103 | 0.44 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr19_-_14064114 | 0.44 |
ENST00000585607.1 ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1 |
podocan-like 1 |
| chr9_+_125132803 | 0.43 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_109935819 | 0.38 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr3_+_12329397 | 0.38 |
ENST00000397015.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr7_+_150020329 | 0.38 |
ENST00000323078.7 |
LRRC61 |
leucine rich repeat containing 61 |
| chr14_+_50234309 | 0.36 |
ENST00000298307.5 |
KLHDC2 |
kelch domain containing 2 |
| chr3_-_149095652 | 0.36 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr1_-_153518270 | 0.34 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr7_+_150020363 | 0.32 |
ENST00000359623.4 ENST00000493307.1 |
LRRC61 |
leucine rich repeat containing 61 |
| chr6_-_32157947 | 0.31 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr17_-_46688334 | 0.31 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr7_+_73868120 | 0.31 |
ENST00000265755.3 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr7_+_73868439 | 0.30 |
ENST00000424337.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr13_-_38172863 | 0.28 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr5_-_176889381 | 0.28 |
ENST00000393563.4 ENST00000512501.1 |
DBN1 |
drebrin 1 |
| chr2_+_90211643 | 0.27 |
ENST00000390277.2 |
IGKV3D-11 |
immunoglobulin kappa variable 3D-11 |
| chr8_-_90769422 | 0.26 |
ENST00000524190.1 ENST00000523859.1 |
RP11-37B2.1 |
RP11-37B2.1 |
| chr2_+_187454749 | 0.25 |
ENST00000261023.3 ENST00000374907.3 |
ITGAV |
integrin, alpha V |
| chr3_+_12329358 | 0.24 |
ENST00000309576.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr2_+_109237717 | 0.24 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr6_+_26440700 | 0.23 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr1_-_101360331 | 0.22 |
ENST00000416479.1 ENST00000370113.3 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
| chr6_-_133035185 | 0.21 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr10_+_101542462 | 0.21 |
ENST00000370449.4 ENST00000370434.1 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr1_-_190446759 | 0.21 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr13_+_53602894 | 0.20 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr9_+_90112590 | 0.20 |
ENST00000472284.1 |
DAPK1 |
death-associated protein kinase 1 |
| chr9_+_90112767 | 0.20 |
ENST00000408954.3 |
DAPK1 |
death-associated protein kinase 1 |
| chr9_-_99540328 | 0.19 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chr2_+_102456277 | 0.19 |
ENST00000421882.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr6_+_42749759 | 0.19 |
ENST00000314073.5 |
GLTSCR1L |
GLTSCR1-like |
| chr16_-_66583701 | 0.19 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr20_+_30697298 | 0.18 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr6_-_52705641 | 0.18 |
ENST00000370989.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr19_+_21324827 | 0.18 |
ENST00000600692.1 ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431 |
zinc finger protein 431 |
| chr20_+_56136136 | 0.17 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr11_-_128894053 | 0.17 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr19_-_14945933 | 0.16 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chr20_+_57594309 | 0.16 |
ENST00000217133.1 |
TUBB1 |
tubulin, beta 1 class VI |
| chr7_+_73868220 | 0.16 |
ENST00000455841.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr15_-_55489097 | 0.15 |
ENST00000260443.4 |
RSL24D1 |
ribosomal L24 domain containing 1 |
| chr19_-_36304201 | 0.14 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr2_+_234826016 | 0.13 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chrX_+_22050546 | 0.13 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr1_-_212965104 | 0.13 |
ENST00000422588.2 ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
| chrX_-_106243451 | 0.13 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chrX_+_77166172 | 0.13 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr16_-_71842941 | 0.13 |
ENST00000423132.2 ENST00000433195.2 ENST00000569748.1 ENST00000570017.1 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
| chr6_+_26365443 | 0.13 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr12_-_22063787 | 0.13 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_-_153919128 | 0.13 |
ENST00000361217.4 |
DENND4B |
DENN/MADD domain containing 4B |
| chr17_-_72772462 | 0.12 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr17_-_53800217 | 0.12 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr2_-_216946500 | 0.12 |
ENST00000265322.7 |
PECR |
peroxisomal trans-2-enoyl-CoA reductase |
| chr15_+_84115868 | 0.12 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr2_-_27886460 | 0.12 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr12_-_23737534 | 0.11 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr11_+_59824127 | 0.11 |
ENST00000278865.3 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr2_-_8977714 | 0.11 |
ENST00000319688.5 ENST00000489024.1 ENST00000256707.3 ENST00000427284.1 ENST00000418530.1 ENST00000473731.1 |
KIDINS220 |
kinase D-interacting substrate, 220kDa |
| chr12_+_104337515 | 0.11 |
ENST00000550595.1 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
| chr18_-_53177984 | 0.11 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr17_+_48823896 | 0.11 |
ENST00000511974.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr6_+_158733692 | 0.11 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr2_-_27886676 | 0.10 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr2_-_21022818 | 0.10 |
ENST00000440866.2 ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43 |
chromosome 2 open reading frame 43 |
| chr6_-_91296602 | 0.10 |
ENST00000369325.3 ENST00000369327.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr9_+_34652164 | 0.10 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr3_+_44840679 | 0.10 |
ENST00000425755.1 |
KIF15 |
kinesin family member 15 |
| chr19_+_49199209 | 0.09 |
ENST00000522966.1 ENST00000425340.2 ENST00000391876.4 |
FUT2 |
fucosyltransferase 2 (secretor status included) |
| chr6_-_91296737 | 0.09 |
ENST00000369332.3 ENST00000369329.3 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
| chr7_-_101272576 | 0.09 |
ENST00000223167.4 |
MYL10 |
myosin, light chain 10, regulatory |
| chr16_-_71843047 | 0.09 |
ENST00000299980.4 ENST00000393512.3 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
| chr11_+_7618413 | 0.09 |
ENST00000528883.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr16_+_28565230 | 0.09 |
ENST00000317058.3 |
CCDC101 |
coiled-coil domain containing 101 |
| chr13_-_36050819 | 0.08 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr2_+_202047596 | 0.08 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr6_+_26402465 | 0.08 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr6_+_26087509 | 0.08 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr3_+_149191723 | 0.08 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr18_-_51750948 | 0.08 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr7_-_140482926 | 0.08 |
ENST00000496384.2 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
| chr8_-_86253888 | 0.08 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr11_+_59824060 | 0.08 |
ENST00000395032.2 ENST00000358152.2 |
MS4A3 |
membrane-spanning 4-domains, subfamily A, member 3 (hematopoietic cell-specific) |
| chr2_-_191115229 | 0.07 |
ENST00000409820.2 ENST00000410045.1 |
HIBCH |
3-hydroxyisobutyryl-CoA hydrolase |
| chr2_-_89385283 | 0.07 |
ENST00000390252.2 |
IGKV3-15 |
immunoglobulin kappa variable 3-15 |
| chr4_+_5527117 | 0.07 |
ENST00000505296.1 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr17_-_39093672 | 0.07 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr2_+_27886330 | 0.07 |
ENST00000326019.6 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr3_+_69812701 | 0.07 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr12_-_25150373 | 0.06 |
ENST00000549828.1 |
C12orf77 |
chromosome 12 open reading frame 77 |
| chr10_+_18549645 | 0.06 |
ENST00000396576.2 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr12_-_10022735 | 0.06 |
ENST00000228438.2 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr18_+_46065393 | 0.06 |
ENST00000256413.3 |
CTIF |
CBP80/20-dependent translation initiation factor |
| chr5_+_175288631 | 0.06 |
ENST00000509837.1 |
CPLX2 |
complexin 2 |
| chr6_-_29527702 | 0.06 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr12_-_112123524 | 0.06 |
ENST00000327551.6 |
BRAP |
BRCA1 associated protein |
| chr6_+_26087646 | 0.05 |
ENST00000309234.6 |
HFE |
hemochromatosis |
| chr17_+_66511540 | 0.05 |
ENST00000588188.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr4_+_37455536 | 0.05 |
ENST00000381980.4 ENST00000508175.1 |
C4orf19 |
chromosome 4 open reading frame 19 |
| chrX_-_100872911 | 0.05 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr5_-_20575959 | 0.05 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr5_-_125930929 | 0.05 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr19_-_3772209 | 0.05 |
ENST00000555978.1 ENST00000555633.1 |
RAX2 |
retina and anterior neural fold homeobox 2 |
| chr2_-_37068530 | 0.05 |
ENST00000593798.1 |
AC007382.1 |
Uncharacterized protein |
| chr2_+_202047843 | 0.05 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr19_-_19887185 | 0.05 |
ENST00000590092.1 ENST00000589910.1 ENST00000589508.1 ENST00000586645.1 ENST00000586816.1 ENST00000588223.1 ENST00000586885.1 |
LINC00663 |
long intergenic non-protein coding RNA 663 |
| chr8_-_57123815 | 0.05 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr12_+_20963632 | 0.05 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chrX_+_108779004 | 0.04 |
ENST00000218004.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr17_+_48823975 | 0.04 |
ENST00000513969.1 ENST00000503728.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr16_-_66584059 | 0.04 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr7_-_64023441 | 0.04 |
ENST00000309683.6 |
ZNF680 |
zinc finger protein 680 |
| chr3_+_141457105 | 0.04 |
ENST00000480908.1 ENST00000393000.3 |
RNF7 |
ring finger protein 7 |
| chr8_+_121137333 | 0.04 |
ENST00000309791.4 ENST00000297848.3 ENST00000247781.3 |
COL14A1 |
collagen, type XIV, alpha 1 |
| chr19_+_21264980 | 0.04 |
ENST00000596053.1 ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714 |
zinc finger protein 714 |
| chr3_+_69812877 | 0.04 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr9_-_21202204 | 0.04 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr1_-_233431458 | 0.04 |
ENST00000258229.9 ENST00000430153.1 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
| chr1_+_10509971 | 0.04 |
ENST00000320498.4 |
CORT |
cortistatin |
| chr3_+_141457030 | 0.04 |
ENST00000273480.3 |
RNF7 |
ring finger protein 7 |
| chr4_+_71108300 | 0.04 |
ENST00000304954.3 |
CSN3 |
casein kappa |
| chr17_-_37607497 | 0.04 |
ENST00000394287.3 ENST00000300651.6 |
MED1 |
mediator complex subunit 1 |
| chr2_+_68962014 | 0.04 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr14_+_102276209 | 0.03 |
ENST00000445439.3 ENST00000334743.5 ENST00000557095.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chrX_+_55744166 | 0.03 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr18_-_52989525 | 0.03 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chrX_+_55744228 | 0.03 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr14_+_72052983 | 0.03 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr12_-_118796910 | 0.03 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr3_+_139063372 | 0.03 |
ENST00000478464.1 |
MRPS22 |
mitochondrial ribosomal protein S22 |
| chr5_-_70316737 | 0.03 |
ENST00000194097.4 |
NAIP |
NLR family, apoptosis inhibitory protein |
| chr6_-_112115074 | 0.03 |
ENST00000368667.2 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr1_+_78769549 | 0.03 |
ENST00000370758.1 |
PTGFR |
prostaglandin F receptor (FP) |
| chr18_-_47018769 | 0.03 |
ENST00000583637.1 ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17 |
ribosomal protein L17 |
| chr3_-_9994021 | 0.03 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chrX_+_134975753 | 0.03 |
ENST00000535938.1 |
SAGE1 |
sarcoma antigen 1 |
| chr5_+_81575281 | 0.03 |
ENST00000380167.4 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr5_-_149829244 | 0.02 |
ENST00000312037.5 |
RPS14 |
ribosomal protein S14 |
| chrX_+_134975858 | 0.02 |
ENST00000537770.1 |
SAGE1 |
sarcoma antigen 1 |
| chr2_-_136678123 | 0.02 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr18_-_52989217 | 0.02 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr15_+_75970672 | 0.02 |
ENST00000435356.1 |
AC105020.1 |
Uncharacterized protein; cDNA FLJ12988 fis, clone NT2RP3000080 |
| chr10_-_115904361 | 0.02 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr1_+_81771806 | 0.02 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr2_-_25391468 | 0.02 |
ENST00000395826.2 |
POMC |
proopiomelanocortin |
| chr2_+_102953608 | 0.02 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr21_-_27423339 | 0.02 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr9_-_21351377 | 0.02 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chr3_+_186353756 | 0.02 |
ENST00000431018.1 ENST00000450521.1 ENST00000539949.1 |
FETUB |
fetuin B |
| chr2_-_25391425 | 0.02 |
ENST00000264708.3 ENST00000449220.1 |
POMC |
proopiomelanocortin |
| chr1_+_57320437 | 0.02 |
ENST00000361249.3 |
C8A |
complement component 8, alpha polypeptide |
| chr4_+_113568207 | 0.02 |
ENST00000511529.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr8_-_30670384 | 0.02 |
ENST00000221138.4 ENST00000518243.1 |
PPP2CB |
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr10_-_101989315 | 0.02 |
ENST00000370397.7 |
CHUK |
conserved helix-loop-helix ubiquitous kinase |
| chr11_+_59856130 | 0.02 |
ENST00000278888.3 |
MS4A2 |
membrane-spanning 4-domains, subfamily A, member 2 |
| chr7_-_44580861 | 0.02 |
ENST00000546276.1 ENST00000289547.4 ENST00000381160.3 ENST00000423141.1 |
NPC1L1 |
NPC1-like 1 |
| chr2_-_74618964 | 0.02 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr11_+_62037622 | 0.02 |
ENST00000227918.2 ENST00000525380.1 |
SCGB2A2 |
secretoglobin, family 2A, member 2 |
| chr1_+_146714291 | 0.02 |
ENST00000431239.1 ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
| chr11_-_13011081 | 0.01 |
ENST00000532541.1 ENST00000526388.1 ENST00000534477.1 ENST00000531402.1 ENST00000527945.1 ENST00000504230.2 |
LINC00958 |
long intergenic non-protein coding RNA 958 |
| chr4_-_100356551 | 0.01 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.0 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.8 | 5.6 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.5 | 1.5 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.4 | 2.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.4 | 1.2 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.3 | 1.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 1.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 0.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 1.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.2 | 0.6 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.8 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.6 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.5 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.9 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 | 2.9 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 1.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.1 | 1.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.9 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.1 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 1.0 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.1 | 0.2 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) response to antineoplastic agent(GO:0097327) |
| 0.1 | 1.0 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.1 | 0.2 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.2 | GO:0043323 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 2.3 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.2 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.4 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.6 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.0 | GO:0072642 | interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.2 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.5 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 1.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 1.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 1.2 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 5.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 1.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.4 | 2.9 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.3 | 1.6 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.3 | 2.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.3 | 1.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.2 | 1.0 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.2 | 1.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.1 | 0.7 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 1.3 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 1.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 1.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.0 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.6 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 1.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.2 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 5.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 1.0 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 2.5 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.8 | GO:0043203 | axon hillock(GO:0043203) |
| 0.3 | 1.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.2 | 0.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 1.7 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 3.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.2 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 2.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 2.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |