Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
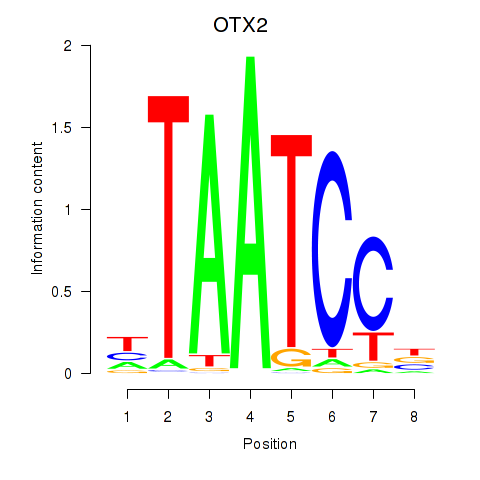
Results for OTX2_CRX
Z-value: 0.47
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | OTX2 |
|
CRX
|
ENSG00000105392.11 | CRX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CRX | hg19_v2_chr19_+_48325097_48325211 | -0.37 | 1.6e-01 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_175711133 | 1.51 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr8_+_70378852 | 0.99 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr5_-_88179302 | 0.98 |
ENST00000504921.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr5_-_88178964 | 0.92 |
ENST00000513252.1 ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr1_+_114522049 | 0.89 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr5_-_88179017 | 0.85 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_+_170366203 | 0.68 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr11_-_111783595 | 0.51 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr14_-_22005018 | 0.47 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr11_-_111794446 | 0.46 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr12_-_91576561 | 0.46 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr16_-_31076332 | 0.44 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chrX_-_6146876 | 0.42 |
ENST00000381095.3 |
NLGN4X |
neuroligin 4, X-linked |
| chr12_-_91576429 | 0.40 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr16_-_31076273 | 0.39 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr1_-_224033596 | 0.39 |
ENST00000391878.2 ENST00000343537.7 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
| chr12_-_91576750 | 0.39 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chrX_-_124097620 | 0.38 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr12_+_81101277 | 0.35 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr5_-_150138061 | 0.35 |
ENST00000521533.1 ENST00000424236.1 |
DCTN4 |
dynactin 4 (p62) |
| chr1_+_26644441 | 0.34 |
ENST00000374213.2 |
CD52 |
CD52 molecule |
| chr14_-_22005062 | 0.34 |
ENST00000317492.5 |
SALL2 |
spalt-like transcription factor 2 |
| chr14_-_22005343 | 0.32 |
ENST00000327430.3 |
SALL2 |
spalt-like transcription factor 2 |
| chrX_-_101771645 | 0.30 |
ENST00000289373.4 |
TMSB15A |
thymosin beta 15a |
| chrX_+_135279179 | 0.29 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr3_+_69915385 | 0.29 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr2_-_175499294 | 0.27 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr12_-_90049828 | 0.26 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chrX_-_15619076 | 0.26 |
ENST00000252519.3 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr19_-_59066452 | 0.26 |
ENST00000312547.2 |
CHMP2A |
charged multivesicular body protein 2A |
| chr12_-_90049878 | 0.24 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chrX_+_135278908 | 0.24 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr18_-_21891460 | 0.24 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr8_-_91095099 | 0.22 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr10_-_76868931 | 0.22 |
ENST00000372700.3 ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13 |
dual specificity phosphatase 13 |
| chr21_-_34915147 | 0.20 |
ENST00000381831.3 ENST00000381839.3 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr2_-_61765315 | 0.19 |
ENST00000406957.1 ENST00000401558.2 |
XPO1 |
exportin 1 (CRM1 homolog, yeast) |
| chr6_-_31697255 | 0.19 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr6_-_31697563 | 0.18 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr1_+_144989309 | 0.18 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr3_-_50649192 | 0.18 |
ENST00000443053.2 ENST00000348721.3 |
CISH |
cytokine inducible SH2-containing protein |
| chr19_-_10121144 | 0.17 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr3_+_4535025 | 0.16 |
ENST00000302640.8 ENST00000354582.6 ENST00000423119.2 ENST00000357086.4 ENST00000456211.2 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
| chr12_-_14849470 | 0.15 |
ENST00000261170.3 |
GUCY2C |
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chrX_+_129473916 | 0.15 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr4_+_129730947 | 0.15 |
ENST00000452328.2 ENST00000504089.1 |
PHF17 |
jade family PHD finger 1 |
| chr14_+_96722152 | 0.15 |
ENST00000216629.6 |
BDKRB1 |
bradykinin receptor B1 |
| chr14_-_71107921 | 0.15 |
ENST00000553982.1 ENST00000500016.1 |
CTD-2540L5.5 CTD-2540L5.6 |
CTD-2540L5.5 CTD-2540L5.6 |
| chr13_-_33780133 | 0.15 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr12_-_16758059 | 0.15 |
ENST00000261169.6 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr17_-_48207157 | 0.14 |
ENST00000330175.4 ENST00000503131.1 |
SAMD14 |
sterile alpha motif domain containing 14 |
| chr1_+_244214577 | 0.14 |
ENST00000358704.4 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
| chr13_+_73632897 | 0.14 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr16_+_81272287 | 0.14 |
ENST00000425577.2 ENST00000564552.1 |
BCMO1 |
beta-carotene 15,15'-monooxygenase 1 |
| chr14_-_101034407 | 0.14 |
ENST00000443071.2 ENST00000557378.1 |
BEGAIN |
brain-enriched guanylate kinase-associated |
| chr18_+_32290218 | 0.13 |
ENST00000348997.5 ENST00000588949.1 ENST00000597599.1 |
DTNA |
dystrobrevin, alpha |
| chr3_+_50649302 | 0.13 |
ENST00000446044.1 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
| chr17_+_43318434 | 0.13 |
ENST00000587489.1 |
FMNL1 |
formin-like 1 |
| chr12_-_16758304 | 0.13 |
ENST00000320122.6 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chrX_+_129473859 | 0.13 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr3_+_49058444 | 0.13 |
ENST00000326925.6 ENST00000395458.2 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr6_-_42690312 | 0.13 |
ENST00000230381.5 |
PRPH2 |
peripherin 2 (retinal degeneration, slow) |
| chr2_-_119605253 | 0.13 |
ENST00000295206.6 |
EN1 |
engrailed homeobox 1 |
| chr1_+_164528866 | 0.12 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr13_-_36050819 | 0.12 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr9_-_130966497 | 0.12 |
ENST00000393608.1 ENST00000372948.3 |
CIZ1 |
CDKN1A interacting zinc finger protein 1 |
| chr7_-_83824169 | 0.12 |
ENST00000265362.4 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr10_+_52751010 | 0.12 |
ENST00000373985.1 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr9_-_95244781 | 0.11 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr10_+_70661014 | 0.11 |
ENST00000373585.3 |
DDX50 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr19_+_56166360 | 0.11 |
ENST00000308924.4 |
U2AF2 |
U2 small nuclear RNA auxiliary factor 2 |
| chr1_-_173174681 | 0.11 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr4_+_129730779 | 0.11 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chr10_+_95848824 | 0.11 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr19_+_2476116 | 0.11 |
ENST00000215631.4 ENST00000587345.1 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
| chr11_-_118436707 | 0.10 |
ENST00000264020.2 ENST00000264021.3 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr11_-_62752162 | 0.10 |
ENST00000458333.2 ENST00000421062.2 |
SLC22A6 |
solute carrier family 22 (organic anion transporter), member 6 |
| chr11_-_75017734 | 0.10 |
ENST00000532525.1 |
ARRB1 |
arrestin, beta 1 |
| chr7_+_90338712 | 0.10 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr7_+_80231466 | 0.10 |
ENST00000309881.7 ENST00000534394.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr12_+_58138664 | 0.10 |
ENST00000257910.3 |
TSPAN31 |
tetraspanin 31 |
| chr10_+_35464513 | 0.10 |
ENST00000494479.1 ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM |
cAMP responsive element modulator |
| chr1_-_150208412 | 0.10 |
ENST00000532744.1 ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr11_-_8680383 | 0.10 |
ENST00000299550.6 |
TRIM66 |
tripartite motif containing 66 |
| chr2_-_225362533 | 0.09 |
ENST00000451538.1 |
CUL3 |
cullin 3 |
| chr3_+_137483579 | 0.09 |
ENST00000306087.1 |
SOX14 |
SRY (sex determining region Y)-box 14 |
| chr11_-_47198380 | 0.09 |
ENST00000419701.2 ENST00000526342.1 ENST00000528444.1 ENST00000530596.1 ENST00000525398.1 ENST00000319543.6 ENST00000426335.2 ENST00000527927.1 ENST00000525314.1 |
ARFGAP2 |
ADP-ribosylation factor GTPase activating protein 2 |
| chr3_-_170626418 | 0.09 |
ENST00000474096.1 ENST00000295822.2 |
EIF5A2 |
eukaryotic translation initiation factor 5A2 |
| chr1_-_241799232 | 0.09 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr1_-_150208291 | 0.09 |
ENST00000533654.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr1_-_150208363 | 0.09 |
ENST00000436748.2 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr12_-_118628350 | 0.09 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr12_+_58138800 | 0.09 |
ENST00000547992.1 ENST00000552816.1 ENST00000547472.1 |
TSPAN31 |
tetraspanin 31 |
| chr10_+_52750930 | 0.08 |
ENST00000401604.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr17_-_46690839 | 0.08 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr12_-_9913489 | 0.08 |
ENST00000228434.3 ENST00000536709.1 |
CD69 |
CD69 molecule |
| chr20_+_42984330 | 0.08 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr19_-_51538148 | 0.08 |
ENST00000319590.4 ENST00000250351.4 |
KLK12 |
kallikrein-related peptidase 12 |
| chr1_-_36615065 | 0.08 |
ENST00000373166.3 ENST00000373159.1 ENST00000373162.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr22_+_41258250 | 0.08 |
ENST00000544094.1 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr3_-_98241358 | 0.08 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr15_+_58430567 | 0.08 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr15_+_58430368 | 0.08 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr3_-_32612263 | 0.08 |
ENST00000432458.2 ENST00000424991.1 ENST00000273130.4 |
DYNC1LI1 |
dynein, cytoplasmic 1, light intermediate chain 1 |
| chr10_-_126716459 | 0.08 |
ENST00000309035.6 |
CTBP2 |
C-terminal binding protein 2 |
| chr1_-_150208498 | 0.07 |
ENST00000314136.8 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr21_+_34915337 | 0.07 |
ENST00000290239.6 ENST00000381692.2 ENST00000300278.4 ENST00000381679.4 |
SON |
SON DNA binding protein |
| chr19_-_51538118 | 0.07 |
ENST00000529888.1 |
KLK12 |
kallikrein-related peptidase 12 |
| chr9_+_82188077 | 0.07 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr18_-_64271363 | 0.07 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr6_-_111804905 | 0.07 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_-_54384425 | 0.07 |
ENST00000375169.3 ENST00000354646.2 |
WNK3 |
WNK lysine deficient protein kinase 3 |
| chr21_-_19775973 | 0.07 |
ENST00000284885.3 |
TMPRSS15 |
transmembrane protease, serine 15 |
| chr19_+_40877583 | 0.07 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chrX_+_100645812 | 0.06 |
ENST00000427805.2 ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A RPL36A-HNRNPH2 |
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr6_+_31555045 | 0.06 |
ENST00000396101.3 ENST00000490742.1 |
LST1 |
leukocyte specific transcript 1 |
| chr12_-_13248562 | 0.06 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chr1_+_202431859 | 0.06 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr6_+_150920999 | 0.06 |
ENST00000367328.1 ENST00000367326.1 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chrX_-_64754611 | 0.05 |
ENST00000374807.5 ENST00000374811.3 ENST00000374804.5 ENST00000312391.8 |
LAS1L |
LAS1-like (S. cerevisiae) |
| chr4_+_70894130 | 0.05 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr2_-_214016314 | 0.05 |
ENST00000434687.1 ENST00000374319.4 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chr8_-_87755878 | 0.05 |
ENST00000320005.5 |
CNGB3 |
cyclic nucleotide gated channel beta 3 |
| chr19_+_39989580 | 0.05 |
ENST00000596614.1 ENST00000205143.4 |
DLL3 |
delta-like 3 (Drosophila) |
| chr22_-_30867973 | 0.04 |
ENST00000402286.1 ENST00000401751.1 ENST00000539629.1 ENST00000403066.1 ENST00000215812.4 |
SEC14L3 |
SEC14-like 3 (S. cerevisiae) |
| chr7_+_136553824 | 0.04 |
ENST00000320658.5 ENST00000453373.1 ENST00000397608.3 ENST00000402486.3 ENST00000401861.1 |
CHRM2 |
cholinergic receptor, muscarinic 2 |
| chr12_-_13248732 | 0.04 |
ENST00000396302.3 |
GSG1 |
germ cell associated 1 |
| chr3_+_49059038 | 0.04 |
ENST00000451378.2 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr1_+_159409512 | 0.04 |
ENST00000423932.3 |
OR10J1 |
olfactory receptor, family 10, subfamily J, member 1 |
| chr6_+_31554962 | 0.04 |
ENST00000376092.3 ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1 |
leukocyte specific transcript 1 |
| chr11_-_118436606 | 0.04 |
ENST00000530872.1 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr5_+_140345820 | 0.04 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr1_-_146082633 | 0.04 |
ENST00000605317.1 ENST00000604938.1 ENST00000339388.5 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr9_-_13279563 | 0.04 |
ENST00000541718.1 |
MPDZ |
multiple PDZ domain protein |
| chr12_-_13248598 | 0.04 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr19_+_39989535 | 0.04 |
ENST00000356433.5 |
DLL3 |
delta-like 3 (Drosophila) |
| chr12_-_13248705 | 0.03 |
ENST00000396310.2 |
GSG1 |
germ cell associated 1 |
| chr6_+_31554636 | 0.03 |
ENST00000433492.1 |
LST1 |
leukocyte specific transcript 1 |
| chrX_+_153409678 | 0.03 |
ENST00000369951.4 |
OPN1LW |
opsin 1 (cone pigments), long-wave-sensitive |
| chr22_+_25615489 | 0.03 |
ENST00000398215.2 |
CRYBB2 |
crystallin, beta B2 |
| chrX_-_153363188 | 0.03 |
ENST00000303391.6 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chr1_+_164529004 | 0.03 |
ENST00000559240.1 ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr13_-_31191642 | 0.03 |
ENST00000405805.1 |
HMGB1 |
high mobility group box 1 |
| chr3_-_45837959 | 0.03 |
ENST00000353278.4 ENST00000456124.2 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chrX_+_69488174 | 0.03 |
ENST00000480877.2 ENST00000307959.8 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr1_+_209757051 | 0.03 |
ENST00000009105.1 ENST00000423146.1 ENST00000361322.2 |
CAMK1G |
calcium/calmodulin-dependent protein kinase IG |
| chr10_+_118350522 | 0.03 |
ENST00000530319.1 ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1 |
pancreatic lipase-related protein 1 |
| chr5_+_150040403 | 0.03 |
ENST00000517768.1 ENST00000297130.4 |
MYOZ3 |
myozenin 3 |
| chr5_-_11588907 | 0.03 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr15_-_35047166 | 0.02 |
ENST00000290374.4 |
GJD2 |
gap junction protein, delta 2, 36kDa |
| chr4_+_68424434 | 0.02 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr3_-_50605150 | 0.02 |
ENST00000357203.3 |
C3orf18 |
chromosome 3 open reading frame 18 |
| chr10_+_118350468 | 0.02 |
ENST00000358834.4 ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1 |
pancreatic lipase-related protein 1 |
| chr6_+_31554826 | 0.02 |
ENST00000376089.2 ENST00000396112.2 |
LST1 |
leukocyte specific transcript 1 |
| chr7_+_136553370 | 0.02 |
ENST00000445907.2 |
CHRM2 |
cholinergic receptor, muscarinic 2 |
| chr3_+_130650738 | 0.02 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr22_-_19137796 | 0.02 |
ENST00000086933.2 |
GSC2 |
goosecoid homeobox 2 |
| chr3_-_45838011 | 0.02 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr19_-_44123734 | 0.02 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr15_+_44092784 | 0.02 |
ENST00000458412.1 |
HYPK |
huntingtin interacting protein K |
| chr10_-_104178857 | 0.02 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr12_-_53171128 | 0.02 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chrX_+_100646190 | 0.02 |
ENST00000471855.1 |
RPL36A |
ribosomal protein L36a |
| chr12_-_86650045 | 0.02 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr10_-_65028938 | 0.02 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr10_+_102505468 | 0.02 |
ENST00000361791.3 ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2 |
paired box 2 |
| chr6_+_42141029 | 0.02 |
ENST00000372958.1 |
GUCA1A |
guanylate cyclase activator 1A (retina) |
| chr14_+_88471468 | 0.02 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr9_+_12693336 | 0.02 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr5_-_33984741 | 0.02 |
ENST00000382102.3 ENST00000509381.1 ENST00000342059.3 ENST00000345083.5 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr3_-_50605077 | 0.02 |
ENST00000426034.1 ENST00000441239.1 |
C3orf18 |
chromosome 3 open reading frame 18 |
| chr9_-_20622478 | 0.02 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_+_162480918 | 0.02 |
ENST00000272716.5 ENST00000446997.1 |
SLC4A10 |
solute carrier family 4, sodium bicarbonate transporter, member 10 |
| chr5_-_33984786 | 0.02 |
ENST00000296589.4 |
SLC45A2 |
solute carrier family 45, member 2 |
| chr19_-_39881669 | 0.02 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr7_-_100171270 | 0.02 |
ENST00000538735.1 |
SAP25 |
Sin3A-associated protein, 25kDa |
| chr12_-_86650077 | 0.02 |
ENST00000552808.2 ENST00000547225.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr16_-_21663919 | 0.02 |
ENST00000569602.1 |
IGSF6 |
immunoglobulin superfamily, member 6 |
| chr15_+_33603147 | 0.01 |
ENST00000415757.3 ENST00000389232.4 |
RYR3 |
ryanodine receptor 3 |
| chr19_-_51537982 | 0.01 |
ENST00000525263.1 |
KLK12 |
kallikrein-related peptidase 12 |
| chr3_+_49057876 | 0.01 |
ENST00000326912.4 |
NDUFAF3 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr3_-_137851220 | 0.01 |
ENST00000236709.3 |
A4GNT |
alpha-1,4-N-acetylglucosaminyltransferase |
| chr5_-_149324306 | 0.01 |
ENST00000255266.5 |
PDE6A |
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr1_+_87380299 | 0.01 |
ENST00000370551.4 ENST00000370550.5 |
HS2ST1 |
heparan sulfate 2-O-sulfotransferase 1 |
| chr11_+_72975559 | 0.01 |
ENST00000349767.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr10_+_104178946 | 0.01 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr18_+_29769978 | 0.01 |
ENST00000269202.6 ENST00000581447.1 |
MEP1B |
meprin A, beta |
| chr1_-_186430222 | 0.01 |
ENST00000391997.2 |
PDC |
phosducin |
| chr1_+_20396649 | 0.01 |
ENST00000375108.3 |
PLA2G5 |
phospholipase A2, group V |
| chr12_-_67197760 | 0.01 |
ENST00000539540.1 ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chrX_-_100129128 | 0.01 |
ENST00000372960.4 ENST00000372964.1 ENST00000217885.5 |
NOX1 |
NADPH oxidase 1 |
| chr11_+_72975578 | 0.01 |
ENST00000393592.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr1_+_159141397 | 0.01 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chrX_+_69488155 | 0.01 |
ENST00000374495.3 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chrX_-_49089771 | 0.01 |
ENST00000376251.1 ENST00000323022.5 ENST00000376265.2 |
CACNA1F |
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr14_-_58894332 | 0.01 |
ENST00000395159.2 |
TIMM9 |
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr13_-_79980315 | 0.01 |
ENST00000438737.2 |
RBM26 |
RNA binding motif protein 26 |
| chr3_-_9291063 | 0.01 |
ENST00000383836.3 |
SRGAP3 |
SLIT-ROBO Rho GTPase activating protein 3 |
| chr12_-_56848426 | 0.01 |
ENST00000257979.4 |
MIP |
major intrinsic protein of lens fiber |
| chr5_-_58882219 | 0.01 |
ENST00000505453.1 ENST00000360047.5 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 1.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 1.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 1.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0003138 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 1.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 1.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.5 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 0.3 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.1 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.1 | 0.6 | GO:0003360 | brainstem development(GO:0003360) |
| 0.1 | 0.1 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 1.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.3 | GO:0003051 | angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.7 | GO:0031272 | regulation of pseudopodium assembly(GO:0031272) |
| 0.0 | 1.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) pyrimidine nucleobase transport(GO:0015855) urea transmembrane transport(GO:0071918) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.4 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 0.1 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 2.8 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.4 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.4 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |