Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
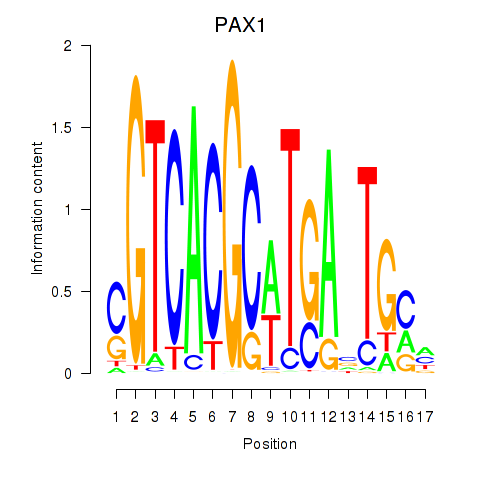
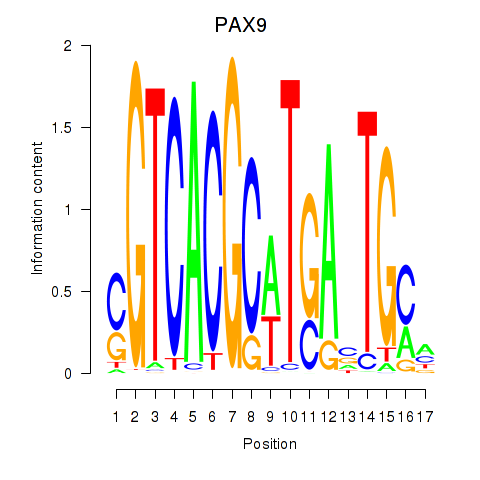
Results for PAX1_PAX9
Z-value: 0.49
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX1
|
ENSG00000125813.9 | PAX1 |
|
PAX9
|
ENSG00000198807.8 | PAX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg19_v2_chr14_+_37126765_37126799, hg19_v2_chr14_+_37131058_37131139 | -0.47 | 6.3e-02 | Click! |
| PAX1 | hg19_v2_chr20_+_21686290_21686311 | -0.39 | 1.4e-01 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23261589 | 1.33 |
ENST00000524168.1 ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2 |
lysyl oxidase-like 2 |
| chr4_+_123747834 | 0.85 |
ENST00000264498.3 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr2_-_175712270 | 0.79 |
ENST00000295497.7 ENST00000444394.1 |
CHN1 |
chimerin 1 |
| chr8_-_101965559 | 0.78 |
ENST00000353245.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr4_+_75230853 | 0.77 |
ENST00000244869.2 |
EREG |
epiregulin |
| chr4_+_123747979 | 0.66 |
ENST00000608478.1 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr18_+_39766626 | 0.66 |
ENST00000593234.1 ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907 |
long intergenic non-protein coding RNA 907 |
| chrX_-_153707545 | 0.63 |
ENST00000357360.4 |
LAGE3 |
L antigen family, member 3 |
| chr9_+_33750515 | 0.58 |
ENST00000361005.5 |
PRSS3 |
protease, serine, 3 |
| chr9_+_33750667 | 0.56 |
ENST00000457896.1 ENST00000342836.4 ENST00000429677.3 |
PRSS3 |
protease, serine, 3 |
| chrX_-_153707246 | 0.55 |
ENST00000407062.1 |
LAGE3 |
L antigen family, member 3 |
| chr8_-_101965146 | 0.49 |
ENST00000395957.2 ENST00000395948.2 ENST00000457309.1 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr9_+_33795533 | 0.43 |
ENST00000379405.3 |
PRSS3 |
protease, serine, 3 |
| chr7_-_19157248 | 0.40 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr17_+_15848231 | 0.39 |
ENST00000304222.2 |
ADORA2B |
adenosine A2b receptor |
| chr9_-_104249400 | 0.29 |
ENST00000374848.3 |
TMEM246 |
transmembrane protein 246 |
| chr8_+_84824920 | 0.28 |
ENST00000523678.1 |
RP11-120I21.2 |
RP11-120I21.2 |
| chr6_+_34857019 | 0.26 |
ENST00000360359.3 ENST00000535627.1 |
ANKS1A |
ankyrin repeat and sterile alpha motif domain containing 1A |
| chr15_+_63569731 | 0.21 |
ENST00000261879.5 |
APH1B |
APH1B gamma secretase subunit |
| chr11_+_6947720 | 0.19 |
ENST00000414517.2 |
ZNF215 |
zinc finger protein 215 |
| chr15_+_63569785 | 0.17 |
ENST00000380343.4 ENST00000560353.1 |
APH1B |
APH1B gamma secretase subunit |
| chrX_-_148669116 | 0.17 |
ENST00000243314.5 |
MAGEA9B |
melanoma antigen family A, 9B |
| chr1_-_213031418 | 0.17 |
ENST00000356684.3 ENST00000426161.1 ENST00000424044.1 |
FLVCR1-AS1 |
FLVCR1 antisense RNA 1 (head to head) |
| chr13_+_21141270 | 0.16 |
ENST00000319980.6 ENST00000537103.1 ENST00000389373.3 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr6_+_74104471 | 0.16 |
ENST00000370336.4 |
DDX43 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr17_-_39968406 | 0.15 |
ENST00000393928.1 |
LEPREL4 |
leprecan-like 4 |
| chr14_+_24540046 | 0.13 |
ENST00000397016.2 ENST00000537691.1 ENST00000560356.1 ENST00000558450.1 |
CPNE6 |
copine VI (neuronal) |
| chr6_-_132945414 | 0.12 |
ENST00000367931.1 ENST00000537809.1 |
TAAR2 |
trace amine associated receptor 2 |
| chr9_-_21142144 | 0.12 |
ENST00000380229.2 |
IFNW1 |
interferon, omega 1 |
| chr13_+_21141208 | 0.12 |
ENST00000351808.5 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
| chrX_+_148622513 | 0.11 |
ENST00000393985.3 ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr1_+_26758790 | 0.11 |
ENST00000427245.2 ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS |
dehydrodolichyl diphosphate synthase |
| chr7_+_142457315 | 0.11 |
ENST00000486171.1 ENST00000311737.7 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr11_+_6947647 | 0.10 |
ENST00000278319.5 |
ZNF215 |
zinc finger protein 215 |
| chr19_-_40791302 | 0.10 |
ENST00000392038.2 ENST00000578123.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr3_+_15247686 | 0.08 |
ENST00000253693.2 |
CAPN7 |
calpain 7 |
| chr19_-_40791211 | 0.07 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr3_+_127771212 | 0.06 |
ENST00000243253.3 ENST00000481210.1 |
SEC61A1 |
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr12_-_101801505 | 0.06 |
ENST00000539055.1 ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1 |
ADP-ribosylation factor-like 1 |
| chr12_+_57828521 | 0.05 |
ENST00000309668.2 |
INHBC |
inhibin, beta C |
| chr10_-_99393242 | 0.04 |
ENST00000370635.3 ENST00000478953.1 ENST00000335628.3 |
MORN4 |
MORN repeat containing 4 |
| chr17_-_39968855 | 0.04 |
ENST00000355468.3 ENST00000590496.1 |
LEPREL4 |
leprecan-like 4 |
| chr2_+_119699864 | 0.04 |
ENST00000541757.1 ENST00000412481.1 |
MARCO |
macrophage receptor with collagenous structure |
| chr10_-_43904608 | 0.04 |
ENST00000337970.3 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
| chr22_+_39745930 | 0.03 |
ENST00000318801.4 ENST00000216155.7 ENST00000406293.3 ENST00000328933.5 |
SYNGR1 |
synaptogyrin 1 |
| chr19_-_5838768 | 0.03 |
ENST00000527106.1 ENST00000531199.1 ENST00000529165.1 |
FUT6 |
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr14_+_39703112 | 0.03 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr6_-_137366163 | 0.03 |
ENST00000367748.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr10_-_99393208 | 0.01 |
ENST00000307450.6 |
MORN4 |
MORN repeat containing 4 |
| chr4_+_619347 | 0.01 |
ENST00000255622.6 |
PDE6B |
phosphodiesterase 6B, cGMP-specific, rod, beta |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.4 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.4 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.2 | 1.5 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.1 | 0.4 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.4 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.1 | 1.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 1.7 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 1.3 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.8 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.1 | GO:0046465 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 | 1.2 | GO:0008033 | tRNA processing(GO:0008033) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 1.6 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |