Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
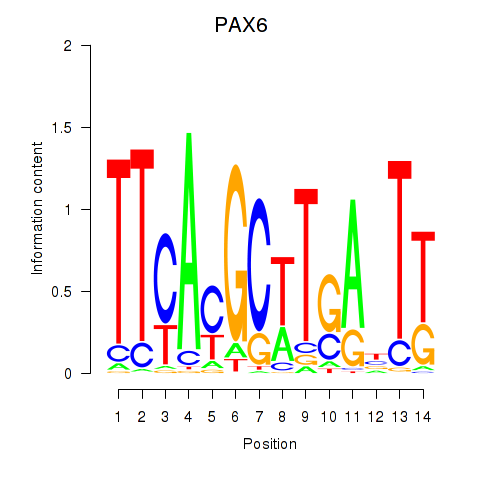
Results for PAX6
Z-value: 1.20
Transcription factors associated with PAX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX6
|
ENSG00000007372.16 | PAX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX6 | hg19_v2_chr11_-_31839488_31839515 | 0.12 | 6.6e-01 | Click! |
Activity profile of PAX6 motif
Sorted Z-values of PAX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_87908600 | 2.87 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chrX_-_124097620 | 2.37 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr6_+_125474795 | 2.14 |
ENST00000304877.13 ENST00000534000.1 ENST00000368402.5 ENST00000368388.2 |
TPD52L1 |
tumor protein D52-like 1 |
| chr6_+_125475335 | 2.03 |
ENST00000532429.1 ENST00000534199.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr6_+_125474939 | 1.98 |
ENST00000527711.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr6_+_125474992 | 1.96 |
ENST00000528193.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr18_+_61420169 | 1.79 |
ENST00000425392.1 ENST00000336429.2 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr18_+_61445007 | 1.69 |
ENST00000447428.1 ENST00000546027.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr12_-_15103621 | 1.56 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chrX_+_37545012 | 1.54 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr6_-_41039567 | 1.38 |
ENST00000468811.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr7_+_50344289 | 1.24 |
ENST00000413698.1 ENST00000359197.5 ENST00000331340.3 ENST00000357364.4 ENST00000343574.5 ENST00000349824.4 ENST00000346667.4 ENST00000440768.2 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr15_+_51973550 | 1.20 |
ENST00000220478.3 |
SCG3 |
secretogranin III |
| chr11_-_58343319 | 1.16 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr15_+_51973680 | 1.15 |
ENST00000542355.2 |
SCG3 |
secretogranin III |
| chr12_-_85306594 | 1.08 |
ENST00000266682.5 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chrX_-_106449656 | 1.00 |
ENST00000372466.4 ENST00000421752.1 ENST00000372461.3 |
NUP62CL |
nucleoporin 62kDa C-terminal like |
| chr6_-_31846744 | 1.00 |
ENST00000414427.1 ENST00000229729.6 ENST00000375562.4 |
SLC44A4 |
solute carrier family 44, member 4 |
| chr6_-_10412600 | 0.99 |
ENST00000379608.3 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr8_-_95487331 | 0.99 |
ENST00000336148.5 |
RAD54B |
RAD54 homolog B (S. cerevisiae) |
| chr9_+_134065506 | 0.94 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr6_+_26156551 | 0.93 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr3_-_119379719 | 0.92 |
ENST00000493094.1 |
POPDC2 |
popeye domain containing 2 |
| chr16_+_84801852 | 0.92 |
ENST00000569925.1 ENST00000567526.1 |
USP10 |
ubiquitin specific peptidase 10 |
| chrX_-_53461288 | 0.90 |
ENST00000375298.4 ENST00000375304.5 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr11_-_28129656 | 0.86 |
ENST00000263181.6 |
KIF18A |
kinesin family member 18A |
| chr9_+_93589734 | 0.86 |
ENST00000375746.1 |
SYK |
spleen tyrosine kinase |
| chr19_-_41903161 | 0.85 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr8_+_142402089 | 0.82 |
ENST00000521578.1 ENST00000520105.1 ENST00000523147.1 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
| chr11_+_60223225 | 0.81 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr11_+_60223312 | 0.81 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr7_-_150754935 | 0.79 |
ENST00000297518.4 |
CDK5 |
cyclin-dependent kinase 5 |
| chrX_-_53461305 | 0.77 |
ENST00000168216.6 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr5_-_169725231 | 0.74 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr2_-_85645545 | 0.71 |
ENST00000409275.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr12_+_113354341 | 0.70 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr21_+_39628655 | 0.69 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr16_-_3767551 | 0.69 |
ENST00000246957.5 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr11_-_11374904 | 0.68 |
ENST00000528848.2 |
CSNK2A3 |
casein kinase 2, alpha 3 polypeptide |
| chr10_-_98031265 | 0.67 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr3_-_124839648 | 0.67 |
ENST00000430155.2 |
SLC12A8 |
solute carrier family 12, member 8 |
| chr21_+_39628852 | 0.65 |
ENST00000398938.2 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr2_-_85625857 | 0.64 |
ENST00000453973.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr2_+_191334212 | 0.63 |
ENST00000444317.1 ENST00000535751.1 |
MFSD6 |
major facilitator superfamily domain containing 6 |
| chr16_-_3767506 | 0.61 |
ENST00000538171.1 |
TRAP1 |
TNF receptor-associated protein 1 |
| chr1_-_201368707 | 0.61 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr1_+_2005425 | 0.60 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr1_+_159409512 | 0.58 |
ENST00000423932.3 |
OR10J1 |
olfactory receptor, family 10, subfamily J, member 1 |
| chr14_+_24099318 | 0.57 |
ENST00000432832.2 |
DHRS2 |
dehydrogenase/reductase (SDR family) member 2 |
| chr7_-_150864635 | 0.56 |
ENST00000297537.4 |
GBX1 |
gastrulation brain homeobox 1 |
| chr17_-_39023462 | 0.56 |
ENST00000251643.4 |
KRT12 |
keratin 12 |
| chr2_-_161056762 | 0.55 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr2_+_114195268 | 0.55 |
ENST00000259199.4 ENST00000416503.2 ENST00000433343.2 |
CBWD2 |
COBW domain containing 2 |
| chr2_-_161056802 | 0.55 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr4_+_87928140 | 0.54 |
ENST00000307808.6 |
AFF1 |
AF4/FMR2 family, member 1 |
| chr19_+_35820064 | 0.54 |
ENST00000341773.6 ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22 |
CD22 molecule |
| chr4_+_115519577 | 0.54 |
ENST00000310836.6 |
UGT8 |
UDP glycosyltransferase 8 |
| chr1_-_201368653 | 0.54 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr1_+_167298281 | 0.53 |
ENST00000367862.5 |
POU2F1 |
POU class 2 homeobox 1 |
| chr2_-_153573965 | 0.52 |
ENST00000448428.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr1_+_77333117 | 0.52 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr2_+_172778952 | 0.51 |
ENST00000392584.1 ENST00000264108.4 |
HAT1 |
histone acetyltransferase 1 |
| chr8_-_95487272 | 0.50 |
ENST00000297592.5 |
RAD54B |
RAD54 homolog B (S. cerevisiae) |
| chr9_-_128246769 | 0.50 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr20_-_32308028 | 0.50 |
ENST00000409299.3 ENST00000217398.3 ENST00000344022.3 |
PXMP4 |
peroxisomal membrane protein 4, 24kDa |
| chr9_+_42704004 | 0.50 |
ENST00000457288.1 |
CBWD7 |
COBW domain containing 7 |
| chr3_-_195997410 | 0.49 |
ENST00000419333.1 |
PCYT1A |
phosphate cytidylyltransferase 1, choline, alpha |
| chr15_+_52155001 | 0.49 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr1_+_87380299 | 0.48 |
ENST00000370551.4 ENST00000370550.5 |
HS2ST1 |
heparan sulfate 2-O-sulfotransferase 1 |
| chr18_+_18822185 | 0.48 |
ENST00000424526.1 ENST00000400483.4 ENST00000431264.1 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr10_-_118765081 | 0.47 |
ENST00000392903.2 ENST00000355371.4 |
KIAA1598 |
KIAA1598 |
| chr4_+_113970772 | 0.45 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr12_-_6716569 | 0.45 |
ENST00000544040.1 ENST00000545942.1 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
| chr8_-_121457608 | 0.44 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr1_+_147374915 | 0.44 |
ENST00000240986.4 |
GJA8 |
gap junction protein, alpha 8, 50kDa |
| chr1_+_174670143 | 0.44 |
ENST00000367687.1 ENST00000347255.2 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr16_-_1401799 | 0.44 |
ENST00000007390.2 |
TSR3 |
TSR3, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr6_-_43027105 | 0.44 |
ENST00000230413.5 ENST00000487429.1 ENST00000489623.1 ENST00000468957.1 |
MRPL2 |
mitochondrial ribosomal protein L2 |
| chr12_+_69080734 | 0.43 |
ENST00000378905.2 |
NUP107 |
nucleoporin 107kDa |
| chr1_-_184006829 | 0.42 |
ENST00000361927.4 |
COLGALT2 |
collagen beta(1-O)galactosyltransferase 2 |
| chr5_-_58882219 | 0.41 |
ENST00000505453.1 ENST00000360047.5 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr3_-_196910721 | 0.41 |
ENST00000443183.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr3_+_15045419 | 0.41 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr8_-_121457332 | 0.41 |
ENST00000518918.1 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr18_+_18822216 | 0.39 |
ENST00000269218.6 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr7_-_76255444 | 0.39 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr10_-_118764862 | 0.37 |
ENST00000260777.10 |
KIAA1598 |
KIAA1598 |
| chr16_-_87350970 | 0.37 |
ENST00000567970.1 |
C16orf95 |
chromosome 16 open reading frame 95 |
| chr2_+_168675182 | 0.36 |
ENST00000305861.1 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr7_+_66461798 | 0.36 |
ENST00000359626.5 ENST00000442959.1 |
TYW1 |
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chrX_+_77154935 | 0.35 |
ENST00000481445.1 |
COX7B |
cytochrome c oxidase subunit VIIb |
| chr1_-_156722015 | 0.35 |
ENST00000368209.5 |
HDGF |
hepatoma-derived growth factor |
| chr9_-_128412696 | 0.34 |
ENST00000420643.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr12_+_6949964 | 0.33 |
ENST00000541978.1 ENST00000435982.2 |
GNB3 |
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr7_-_97501706 | 0.33 |
ENST00000455086.1 ENST00000453600.1 |
ASNS |
asparagine synthetase (glutamine-hydrolyzing) |
| chr5_+_85913721 | 0.32 |
ENST00000247655.3 ENST00000509578.1 ENST00000515763.1 |
COX7C |
cytochrome c oxidase subunit VIIc |
| chr18_-_45663666 | 0.32 |
ENST00000535628.2 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
| chr5_-_98262240 | 0.31 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr2_-_29093132 | 0.31 |
ENST00000306108.5 |
TRMT61B |
tRNA methyltransferase 61 homolog B (S. cerevisiae) |
| chr3_-_101232019 | 0.31 |
ENST00000394095.2 ENST00000394091.1 ENST00000394094.2 ENST00000358203.3 ENST00000348610.3 ENST00000314261.7 |
SENP7 |
SUMO1/sentrin specific peptidase 7 |
| chr6_+_88299833 | 0.30 |
ENST00000392844.3 ENST00000257789.4 ENST00000546266.1 ENST00000417380.2 |
ORC3 |
origin recognition complex, subunit 3 |
| chr16_+_84682108 | 0.30 |
ENST00000564996.1 ENST00000258157.5 ENST00000567410.1 |
KLHL36 |
kelch-like family member 36 |
| chr18_+_13611431 | 0.29 |
ENST00000587757.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr7_-_97501733 | 0.29 |
ENST00000444334.1 ENST00000422745.1 ENST00000394308.3 ENST00000451771.1 ENST00000175506.4 |
ASNS |
asparagine synthetase (glutamine-hydrolyzing) |
| chr10_+_97471508 | 0.27 |
ENST00000453258.2 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr19_+_50936142 | 0.26 |
ENST00000357701.5 |
MYBPC2 |
myosin binding protein C, fast type |
| chr3_-_15563229 | 0.26 |
ENST00000383786.5 ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr5_+_140254884 | 0.26 |
ENST00000398631.2 |
PCDHA12 |
protocadherin alpha 12 |
| chr19_+_15852203 | 0.25 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr8_-_21771214 | 0.25 |
ENST00000276420.4 |
DOK2 |
docking protein 2, 56kDa |
| chr17_-_2415169 | 0.25 |
ENST00000263092.6 ENST00000538844.1 ENST00000576976.1 |
METTL16 |
methyltransferase like 16 |
| chr6_-_64029879 | 0.25 |
ENST00000370658.5 ENST00000485906.2 ENST00000370657.4 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
| chr20_+_48807351 | 0.25 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr8_-_33370607 | 0.24 |
ENST00000360742.5 ENST00000523305.1 |
TTI2 |
TELO2 interacting protein 2 |
| chr10_+_102222798 | 0.24 |
ENST00000343737.5 |
WNT8B |
wingless-type MMTV integration site family, member 8B |
| chr2_-_154335300 | 0.24 |
ENST00000325926.3 |
RPRM |
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr16_-_51185172 | 0.23 |
ENST00000251020.4 |
SALL1 |
spalt-like transcription factor 1 |
| chr12_+_10460549 | 0.23 |
ENST00000543420.1 ENST00000543777.1 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr1_-_86174065 | 0.23 |
ENST00000370574.3 ENST00000431532.2 |
ZNHIT6 |
zinc finger, HIT-type containing 6 |
| chrX_-_65259914 | 0.23 |
ENST00000374737.4 ENST00000455586.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr3_+_171561127 | 0.23 |
ENST00000334567.5 ENST00000450693.1 |
TMEM212 |
transmembrane protein 212 |
| chr17_+_7239821 | 0.23 |
ENST00000158762.3 ENST00000570457.2 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr17_-_73389737 | 0.22 |
ENST00000392563.1 |
GRB2 |
growth factor receptor-bound protein 2 |
| chr17_+_18380051 | 0.22 |
ENST00000581545.1 ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C |
lectin, galactoside-binding, soluble, 9C |
| chr2_+_220363579 | 0.22 |
ENST00000313597.5 ENST00000373917.3 ENST00000358215.3 ENST00000373908.1 ENST00000455657.1 ENST00000435316.1 ENST00000341142.3 |
GMPPA |
GDP-mannose pyrophosphorylase A |
| chr4_-_87281224 | 0.22 |
ENST00000395169.3 ENST00000395161.2 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr1_+_180897269 | 0.21 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chr3_-_142297668 | 0.21 |
ENST00000350721.4 ENST00000383101.3 |
ATR |
ataxia telangiectasia and Rad3 related |
| chr17_-_79633590 | 0.21 |
ENST00000374741.3 ENST00000571503.1 |
OXLD1 |
oxidoreductase-like domain containing 1 |
| chr1_-_165738072 | 0.21 |
ENST00000481278.1 |
TMCO1 |
transmembrane and coiled-coil domains 1 |
| chr17_-_58096336 | 0.20 |
ENST00000587125.1 ENST00000407042.3 |
TBC1D3P1-DHX40P1 |
TBC1D3P1-DHX40P1 readthrough transcribed pseudogene |
| chr1_-_229644034 | 0.20 |
ENST00000366678.3 ENST00000261396.3 ENST00000537506.1 |
NUP133 |
nucleoporin 133kDa |
| chr6_-_33041378 | 0.19 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr1_+_174669653 | 0.19 |
ENST00000325589.5 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chrX_-_80457385 | 0.18 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr10_+_91092241 | 0.18 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr11_-_102576537 | 0.18 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr2_+_232135245 | 0.17 |
ENST00000446447.1 |
ARMC9 |
armadillo repeat containing 9 |
| chr5_-_135231516 | 0.16 |
ENST00000274520.1 |
IL9 |
interleukin 9 |
| chr10_+_13628933 | 0.16 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr5_-_55529115 | 0.16 |
ENST00000513241.2 ENST00000341048.4 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr4_-_88141755 | 0.16 |
ENST00000273963.5 |
KLHL8 |
kelch-like family member 8 |
| chr14_+_88471468 | 0.16 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr19_+_51628165 | 0.16 |
ENST00000250360.3 ENST00000440804.3 |
SIGLEC9 |
sialic acid binding Ig-like lectin 9 |
| chr9_-_95166884 | 0.16 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr15_-_23034322 | 0.16 |
ENST00000539711.2 ENST00000560039.1 ENST00000398013.3 ENST00000337451.3 ENST00000359727.4 ENST00000398014.2 |
NIPA2 |
non imprinted in Prader-Willi/Angelman syndrome 2 |
| chr12_-_45270077 | 0.15 |
ENST00000551601.1 ENST00000549027.1 ENST00000452445.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr13_+_96085847 | 0.15 |
ENST00000376873.3 |
CLDN10 |
claudin 10 |
| chr12_-_45270151 | 0.15 |
ENST00000429094.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr16_-_71264558 | 0.15 |
ENST00000448089.2 ENST00000393550.2 ENST00000448691.1 ENST00000393567.2 ENST00000321489.5 ENST00000539973.1 ENST00000288168.10 ENST00000545267.1 ENST00000541601.1 ENST00000538248.1 |
HYDIN |
HYDIN, axonemal central pair apparatus protein |
| chr17_+_25958174 | 0.15 |
ENST00000313648.6 ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9 |
lectin, galactoside-binding, soluble, 9 |
| chr4_+_187148556 | 0.15 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr11_-_117747434 | 0.15 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr11_-_57298187 | 0.14 |
ENST00000525158.1 ENST00000257245.4 ENST00000525587.1 |
TIMM10 |
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr12_-_10151773 | 0.14 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr16_-_18911366 | 0.14 |
ENST00000565224.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr8_+_59323823 | 0.13 |
ENST00000399598.2 |
UBXN2B |
UBX domain protein 2B |
| chr12_-_56352368 | 0.13 |
ENST00000549404.1 |
PMEL |
premelanosome protein |
| chr16_+_31128978 | 0.13 |
ENST00000448516.2 ENST00000219797.4 |
KAT8 |
K(lysine) acetyltransferase 8 |
| chr12_-_45269430 | 0.12 |
ENST00000395487.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr19_+_16059818 | 0.12 |
ENST00000322107.1 |
OR10H4 |
olfactory receptor, family 10, subfamily H, member 4 |
| chr9_-_73736511 | 0.12 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr12_-_47473425 | 0.11 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr7_+_65552756 | 0.11 |
ENST00000450043.1 |
AC068533.7 |
AC068533.7 |
| chr19_-_50370509 | 0.11 |
ENST00000596014.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chr11_+_14926543 | 0.11 |
ENST00000523376.1 |
CALCB |
calcitonin-related polypeptide beta |
| chrX_-_55024967 | 0.11 |
ENST00000545676.1 |
PFKFB1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr3_-_195310802 | 0.11 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr7_-_152373216 | 0.11 |
ENST00000359321.1 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr12_+_27175476 | 0.11 |
ENST00000546323.1 ENST00000282892.3 |
MED21 |
mediator complex subunit 21 |
| chr2_+_202098166 | 0.11 |
ENST00000392263.2 ENST00000264274.9 ENST00000392259.2 ENST00000392266.3 ENST00000432109.2 ENST00000264275.5 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
| chr11_-_117747327 | 0.11 |
ENST00000584230.1 ENST00000527429.1 ENST00000584394.1 ENST00000532984.1 |
FXYD6 FXYD6-FXYD2 |
FXYD domain containing ion transport regulator 6 FXYD6-FXYD2 readthrough |
| chr8_-_21771182 | 0.10 |
ENST00000523932.1 ENST00000544659.1 |
DOK2 |
docking protein 2, 56kDa |
| chr4_-_68749745 | 0.10 |
ENST00000283916.6 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr18_+_13611763 | 0.09 |
ENST00000585931.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr2_-_163008903 | 0.09 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr1_-_89458415 | 0.08 |
ENST00000321792.5 ENST00000370491.3 |
RBMXL1 CCBL2 |
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr15_+_52043758 | 0.08 |
ENST00000249700.4 ENST00000539962.2 |
TMOD2 |
tropomodulin 2 (neuronal) |
| chr18_+_32173276 | 0.08 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr12_-_7848364 | 0.08 |
ENST00000329913.3 |
GDF3 |
growth differentiation factor 3 |
| chr1_+_39491984 | 0.07 |
ENST00000372969.3 ENST00000372967.3 |
NDUFS5 |
NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) |
| chr20_-_44144249 | 0.07 |
ENST00000217428.6 |
SPINT3 |
serine peptidase inhibitor, Kunitz type, 3 |
| chr1_+_26758790 | 0.07 |
ENST00000427245.2 ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS |
dehydrodolichyl diphosphate synthase |
| chr5_+_140602904 | 0.07 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr10_-_17243579 | 0.07 |
ENST00000525762.1 ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
| chr2_-_86116093 | 0.07 |
ENST00000377332.3 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr12_+_10460417 | 0.07 |
ENST00000381908.3 ENST00000336164.4 ENST00000350274.5 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr2_-_70520539 | 0.07 |
ENST00000482975.2 ENST00000438261.1 |
SNRPG |
small nuclear ribonucleoprotein polypeptide G |
| chr2_+_29320571 | 0.07 |
ENST00000401605.1 ENST00000401617.2 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr10_-_27389320 | 0.06 |
ENST00000436985.2 |
ANKRD26 |
ankyrin repeat domain 26 |
| chr4_-_68749699 | 0.06 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr4_+_76995855 | 0.06 |
ENST00000355810.4 ENST00000349321.3 |
ART3 |
ADP-ribosyltransferase 3 |
| chr7_+_76109827 | 0.06 |
ENST00000446820.2 |
DTX2 |
deltex homolog 2 (Drosophila) |
| chr1_+_79115503 | 0.06 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr12_-_121476959 | 0.05 |
ENST00000339275.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr17_+_43224684 | 0.05 |
ENST00000332499.2 |
HEXIM1 |
hexamethylene bis-acetamide inducible 1 |
| chr15_-_65282274 | 0.05 |
ENST00000204566.2 |
SPG21 |
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr16_+_4897912 | 0.05 |
ENST00000545171.1 |
UBN1 |
ubinuclein 1 |
| chr3_-_15540055 | 0.04 |
ENST00000605797.1 ENST00000435459.2 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr2_+_145780739 | 0.04 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 3.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.4 | 1.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 1.7 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.3 | 0.9 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 0.8 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.7 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.3 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.1 | 0.3 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.1 | 1.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.3 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.1 | 1.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.9 | GO:1990023 | kinetochore microtubule(GO:0005828) mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.4 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 1.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 2.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 1.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.9 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.2 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.3 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 2.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.5 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.7 | 2.0 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.5 | 1.5 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.4 | 2.9 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.4 | 1.6 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 1.0 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.3 | 1.3 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.3 | 0.9 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.0 | GO:0003409 | optic cup structural organization(GO:0003409) |
| 0.2 | 1.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.2 | 0.9 | GO:0042223 | response to molecule of fungal origin(GO:0002238) regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.2 | 0.6 | GO:0070981 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.2 | 0.5 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.2 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.1 | 0.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 0.8 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.7 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.9 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.6 | GO:0019230 | proprioception(GO:0019230) |
| 0.1 | 1.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.3 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.7 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.2 | GO:0072092 | olfactory bulb mitral cell layer development(GO:0061034) ureteric bud invasion(GO:0072092) |
| 0.1 | 2.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.5 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.1 | 0.8 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.2 | GO:1904884 | telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.1 | 0.9 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.1 | 0.4 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 1.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.3 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.1 | 0.4 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.2 | GO:0071033 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of pre-mRNA at the site of transcription(GO:0071033) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.1 | 1.6 | GO:0006312 | mitotic recombination(GO:0006312) |
| 0.1 | 8.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 0.2 | GO:0031081 | nuclear pore distribution(GO:0031081) nuclear pore localization(GO:0051664) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.5 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.1 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.1 | GO:0039507 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 1.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.6 | GO:0043011 | myeloid dendritic cell differentiation(GO:0043011) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.9 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 1.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.3 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.8 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.3 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.2 | GO:0032754 | positive regulation of interleukin-5 production(GO:0032754) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.5 | 1.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 1.0 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.2 | 1.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.2 | 0.8 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.2 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 1.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.9 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.8 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.6 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.4 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 0.6 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.3 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 2.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.7 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 1.3 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.7 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.1 | 1.0 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.8 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.9 | GO:0035325 | receptor signaling protein tyrosine kinase activity(GO:0004716) Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 1.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.4 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 3.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 2.0 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.7 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.4 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 10.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.9 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |