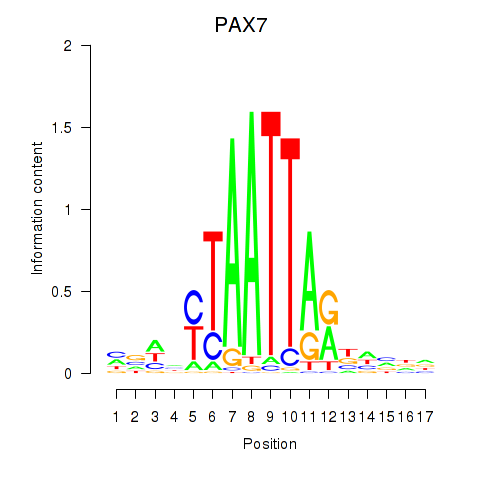
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for PAX7_NOBOX
Z-value: 0.65
Transcription factors associated with PAX7_NOBOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX7
|
ENSG00000009709.7 | PAX7 |
|
NOBOX
|
ENSG00000106410.10 | NOBOX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX7 | hg19_v2_chr1_+_18957500_18957623 | 0.25 | 3.4e-01 | Click! |
Activity profile of PAX7_NOBOX motif
Sorted Z-values of PAX7_NOBOX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX7_NOBOX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102668879 | 1.91 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr3_-_149095652 | 1.30 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr2_+_102953608 | 1.15 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chrX_+_43515467 | 0.97 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr1_+_81771806 | 0.95 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr1_+_226013047 | 0.84 |
ENST00000366837.4 |
EPHX1 |
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr11_-_33913708 | 0.83 |
ENST00000257818.2 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
| chr6_-_112575912 | 0.81 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr12_-_15038779 | 0.73 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr10_+_94451574 | 0.72 |
ENST00000492654.2 |
HHEX |
hematopoietically expressed homeobox |
| chr3_-_160823158 | 0.67 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr14_-_74959994 | 0.61 |
ENST00000238633.2 ENST00000434013.2 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr3_-_160823040 | 0.60 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr14_-_74960030 | 0.60 |
ENST00000553490.1 ENST00000557510.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr6_-_112575687 | 0.60 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr1_-_92371839 | 0.56 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr14_-_74959978 | 0.56 |
ENST00000541064.1 |
NPC2 |
Niemann-Pick disease, type C2 |
| chr6_-_112575838 | 0.53 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr6_-_112575758 | 0.40 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr3_+_148709128 | 0.39 |
ENST00000345003.4 ENST00000296048.6 ENST00000483267.1 |
GYG1 |
glycogenin 1 |
| chr2_-_225266743 | 0.37 |
ENST00000409685.3 |
FAM124B |
family with sequence similarity 124B |
| chr12_-_91546926 | 0.37 |
ENST00000550758.1 |
DCN |
decorin |
| chr16_-_66584059 | 0.36 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr20_+_19867150 | 0.36 |
ENST00000255006.6 |
RIN2 |
Ras and Rab interactor 2 |
| chr7_+_107224364 | 0.35 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr14_-_52535712 | 0.35 |
ENST00000216286.5 ENST00000541773.1 |
NID2 |
nidogen 2 (osteonidogen) |
| chr19_+_50016411 | 0.35 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr12_-_10022735 | 0.35 |
ENST00000228438.2 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr1_+_186798073 | 0.35 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr2_-_74618964 | 0.33 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr6_+_34204642 | 0.31 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr11_+_77532233 | 0.31 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr14_+_104182061 | 0.30 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr4_-_57547454 | 0.30 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr2_+_90108504 | 0.29 |
ENST00000390271.2 |
IGKV6D-41 |
immunoglobulin kappa variable 6D-41 (non-functional) |
| chr2_+_68962014 | 0.28 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr10_+_5135981 | 0.28 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr4_-_57547870 | 0.28 |
ENST00000381260.3 ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX |
HOP homeobox |
| chr14_+_104182105 | 0.26 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr11_+_77532155 | 0.26 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr16_-_66583701 | 0.26 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr6_-_110501200 | 0.26 |
ENST00000392586.1 ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1 |
WAS protein family, member 1 |
| chr2_-_74619152 | 0.25 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr7_+_115862858 | 0.25 |
ENST00000393481.2 |
TES |
testis derived transcript (3 LIM domains) |
| chr15_+_58724184 | 0.25 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr1_+_12916941 | 0.24 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr21_+_35014783 | 0.22 |
ENST00000381291.4 ENST00000381285.4 ENST00000399367.3 ENST00000399352.1 ENST00000399355.2 ENST00000399349.1 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
| chr4_+_186990298 | 0.21 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chr6_-_167040731 | 0.20 |
ENST00000265678.4 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr10_-_131909071 | 0.20 |
ENST00000456581.1 |
LINC00959 |
long intergenic non-protein coding RNA 959 |
| chr5_+_140593509 | 0.19 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chr6_-_32145861 | 0.19 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr19_+_50016610 | 0.19 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr15_-_37393406 | 0.19 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr18_-_21977748 | 0.19 |
ENST00000399441.4 ENST00000319481.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr4_+_74275057 | 0.19 |
ENST00000511370.1 |
ALB |
albumin |
| chr7_+_138145076 | 0.19 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr7_+_134528635 | 0.18 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr15_+_64680003 | 0.18 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr11_-_327537 | 0.17 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr8_+_9413410 | 0.17 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr8_+_105235572 | 0.17 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr18_+_29171689 | 0.17 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr9_+_470288 | 0.17 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr4_-_141348789 | 0.16 |
ENST00000414773.1 |
CLGN |
calmegin |
| chr3_-_46000064 | 0.16 |
ENST00000433878.1 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr1_+_155278625 | 0.15 |
ENST00000368356.4 ENST00000356657.6 |
FDPS |
farnesyl diphosphate synthase |
| chr4_-_83812402 | 0.15 |
ENST00000395310.2 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr4_-_83812248 | 0.15 |
ENST00000514326.1 ENST00000505434.1 ENST00000503058.1 ENST00000348405.4 ENST00000505984.1 ENST00000513858.1 ENST00000508479.1 ENST00000443462.2 ENST00000508502.1 ENST00000509142.1 ENST00000432794.1 ENST00000448323.1 ENST00000326950.5 ENST00000311785.7 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr4_+_156587853 | 0.14 |
ENST00000506455.1 ENST00000511108.1 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr2_+_152214098 | 0.14 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr6_+_26365443 | 0.14 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr19_-_46088068 | 0.14 |
ENST00000263275.4 ENST00000323060.3 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr16_-_55866997 | 0.14 |
ENST00000360526.3 ENST00000361503.4 |
CES1 |
carboxylesterase 1 |
| chr7_-_22234381 | 0.14 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_-_10978957 | 0.14 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr3_-_171528227 | 0.14 |
ENST00000356327.5 ENST00000342215.6 ENST00000340989.4 ENST00000351298.4 |
PLD1 |
phospholipase D1, phosphatidylcholine-specific |
| chr14_-_67878917 | 0.14 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr1_+_155278539 | 0.14 |
ENST00000447866.1 |
FDPS |
farnesyl diphosphate synthase |
| chr8_+_54764346 | 0.13 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chrX_+_95939638 | 0.13 |
ENST00000373061.3 ENST00000373054.4 ENST00000355827.4 |
DIAPH2 |
diaphanous-related formin 2 |
| chr19_-_14606900 | 0.13 |
ENST00000393029.3 ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
| chr3_+_138340049 | 0.13 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr14_-_25078864 | 0.13 |
ENST00000216338.4 ENST00000557220.2 ENST00000382548.4 |
GZMH |
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr1_-_1259989 | 0.13 |
ENST00000540437.1 |
CPSF3L |
cleavage and polyadenylation specific factor 3-like |
| chr20_-_50722183 | 0.13 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr11_+_17298255 | 0.12 |
ENST00000531172.1 ENST00000533738.2 ENST00000323688.6 |
NUCB2 |
nucleobindin 2 |
| chr2_-_27886460 | 0.12 |
ENST00000404798.2 ENST00000405491.1 ENST00000464789.2 ENST00000406540.1 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr20_-_33735070 | 0.12 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr17_+_63133526 | 0.12 |
ENST00000443584.3 |
RGS9 |
regulator of G-protein signaling 9 |
| chrX_-_134156502 | 0.12 |
ENST00000391440.1 |
FAM127C |
family with sequence similarity 127, member C |
| chr11_+_17298297 | 0.12 |
ENST00000529010.1 |
NUCB2 |
nucleobindin 2 |
| chr4_+_88896819 | 0.12 |
ENST00000237623.7 ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1 |
secreted phosphoprotein 1 |
| chr7_-_87342564 | 0.11 |
ENST00000265724.3 ENST00000416177.1 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| chr1_+_1260147 | 0.11 |
ENST00000343938.4 |
GLTPD1 |
glycolipid transfer protein domain containing 1 |
| chr1_-_1260010 | 0.11 |
ENST00000434694.2 ENST00000421495.2 ENST00000545578.1 ENST00000419704.1 ENST00000530031.1 ENST00000526332.1 ENST00000498476.2 ENST00000450926.2 ENST00000527719.1 ENST00000534345.1 ENST00000411962.1 ENST00000435064.1 |
CPSF3L |
cleavage and polyadenylation specific factor 3-like |
| chr12_+_3000037 | 0.11 |
ENST00000544943.1 ENST00000448120.2 |
TULP3 |
tubby like protein 3 |
| chr17_+_7155819 | 0.11 |
ENST00000570322.1 ENST00000576496.1 ENST00000574841.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr2_+_234826016 | 0.11 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr3_+_47324424 | 0.11 |
ENST00000437353.1 ENST00000232766.5 ENST00000455924.2 |
KLHL18 |
kelch-like family member 18 |
| chr20_+_814377 | 0.10 |
ENST00000304189.2 ENST00000381939.1 |
FAM110A |
family with sequence similarity 110, member A |
| chr16_+_68279207 | 0.10 |
ENST00000413021.2 ENST00000565744.1 ENST00000219345.5 |
PLA2G15 |
phospholipase A2, group XV |
| chr10_+_101542462 | 0.10 |
ENST00000370449.4 ENST00000370434.1 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr3_-_108248169 | 0.10 |
ENST00000273353.3 |
MYH15 |
myosin, heavy chain 15 |
| chr11_-_22647350 | 0.10 |
ENST00000327470.3 |
FANCF |
Fanconi anemia, complementation group F |
| chr2_-_27886676 | 0.10 |
ENST00000337768.5 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
| chr6_+_26402517 | 0.10 |
ENST00000414912.2 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr12_-_65146636 | 0.10 |
ENST00000418919.2 |
GNS |
glucosamine (N-acetyl)-6-sulfatase |
| chr2_+_68961934 | 0.10 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr1_-_43855479 | 0.10 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr1_+_12851545 | 0.10 |
ENST00000332296.7 |
PRAMEF1 |
PRAME family member 1 |
| chr6_+_26402465 | 0.09 |
ENST00000476549.2 ENST00000289361.6 ENST00000450085.2 ENST00000425234.2 ENST00000427334.1 ENST00000506698.1 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
| chr1_-_77685084 | 0.09 |
ENST00000370812.3 ENST00000359130.1 ENST00000445065.1 ENST00000370813.5 |
PIGK |
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr4_+_69681710 | 0.09 |
ENST00000265403.7 ENST00000458688.2 |
UGT2B10 |
UDP glucuronosyltransferase 2 family, polypeptide B10 |
| chr19_-_53758094 | 0.09 |
ENST00000601828.1 ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677 |
zinc finger protein 677 |
| chr12_+_3000073 | 0.09 |
ENST00000397132.2 |
TULP3 |
tubby like protein 3 |
| chr17_+_73452695 | 0.09 |
ENST00000582186.1 ENST00000582455.1 ENST00000581252.1 ENST00000579208.1 |
KIAA0195 |
KIAA0195 |
| chr17_+_79650962 | 0.09 |
ENST00000329138.4 |
HGS |
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr6_+_26440700 | 0.09 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr7_+_100210133 | 0.08 |
ENST00000393950.2 ENST00000424091.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chrX_+_52780318 | 0.08 |
ENST00000375515.3 ENST00000276049.6 |
SSX2B |
synovial sarcoma, X breakpoint 2B |
| chr6_+_32146131 | 0.08 |
ENST00000375094.3 |
RNF5 |
ring finger protein 5, E3 ubiquitin protein ligase |
| chr2_+_68961905 | 0.08 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr19_+_11485333 | 0.08 |
ENST00000312423.2 |
SWSAP1 |
SWIM-type zinc finger 7 associated protein 1 |
| chr3_+_152552685 | 0.08 |
ENST00000305097.3 |
P2RY1 |
purinergic receptor P2Y, G-protein coupled, 1 |
| chrX_+_67913471 | 0.08 |
ENST00000374597.3 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
| chr5_+_140227357 | 0.08 |
ENST00000378122.3 |
PCDHA9 |
protocadherin alpha 9 |
| chr1_+_224370873 | 0.08 |
ENST00000323699.4 ENST00000391877.3 |
DEGS1 |
delta(4)-desaturase, sphingolipid 1 |
| chr11_+_67798114 | 0.07 |
ENST00000453471.2 ENST00000528492.1 ENST00000526339.1 ENST00000525419.1 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr17_-_7167279 | 0.07 |
ENST00000571932.2 |
CLDN7 |
claudin 7 |
| chr11_+_67798090 | 0.07 |
ENST00000313468.5 |
NDUFS8 |
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr2_+_145780767 | 0.07 |
ENST00000599358.1 ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr6_+_27100811 | 0.07 |
ENST00000359193.2 |
HIST1H2AG |
histone cluster 1, H2ag |
| chr19_+_48972265 | 0.07 |
ENST00000452733.2 |
CYTH2 |
cytohesin 2 |
| chr7_+_120628731 | 0.07 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr7_-_2883928 | 0.07 |
ENST00000275364.3 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
| chrX_+_48114752 | 0.07 |
ENST00000376919.3 |
SSX1 |
synovial sarcoma, X breakpoint 1 |
| chr1_-_13452656 | 0.07 |
ENST00000376132.3 |
PRAMEF13 |
PRAME family member 13 |
| chr9_-_21202204 | 0.07 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr12_-_23737534 | 0.07 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr11_-_35287243 | 0.07 |
ENST00000464522.2 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_+_156544057 | 0.07 |
ENST00000498839.1 ENST00000470811.1 ENST00000356539.4 ENST00000483177.1 ENST00000477399.1 ENST00000491763.1 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
| chr17_+_72772621 | 0.07 |
ENST00000335464.5 ENST00000417024.2 ENST00000578764.1 ENST00000582773.1 ENST00000582330.1 |
TMEM104 |
transmembrane protein 104 |
| chr19_-_10227503 | 0.07 |
ENST00000593054.1 |
EIF3G |
eukaryotic translation initiation factor 3, subunit G |
| chr8_-_86290333 | 0.07 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr5_+_66300446 | 0.07 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr2_-_89160770 | 0.07 |
ENST00000390240.2 |
IGKJ3 |
immunoglobulin kappa joining 3 |
| chr19_+_48972459 | 0.07 |
ENST00000427476.1 |
CYTH2 |
cytohesin 2 |
| chr5_+_95066823 | 0.07 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr11_+_24518723 | 0.07 |
ENST00000336930.6 ENST00000529015.1 ENST00000533227.1 |
LUZP2 |
leucine zipper protein 2 |
| chr11_-_117748138 | 0.06 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr19_+_41770269 | 0.06 |
ENST00000378215.4 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr5_-_10761206 | 0.06 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr17_-_18266660 | 0.06 |
ENST00000582653.1 ENST00000352886.6 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr3_+_138340067 | 0.06 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chrX_-_153363125 | 0.06 |
ENST00000407218.1 ENST00000453960.2 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chrX_+_95939711 | 0.06 |
ENST00000373049.4 ENST00000324765.8 |
DIAPH2 |
diaphanous-related formin 2 |
| chr6_+_52051171 | 0.06 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chr1_+_151739131 | 0.06 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr11_-_32457176 | 0.06 |
ENST00000332351.3 |
WT1 |
Wilms tumor 1 |
| chr1_-_153919128 | 0.06 |
ENST00000361217.4 |
DENND4B |
DENN/MADD domain containing 4B |
| chr19_-_14064114 | 0.06 |
ENST00000585607.1 ENST00000538517.2 ENST00000587458.1 ENST00000538371.2 |
PODNL1 |
podocan-like 1 |
| chr14_-_94789663 | 0.06 |
ENST00000557225.1 ENST00000341584.3 |
SERPINA6 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr9_-_128003606 | 0.06 |
ENST00000324460.6 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
| chr4_+_169418195 | 0.06 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr2_-_55276320 | 0.06 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chrX_-_48271344 | 0.06 |
ENST00000376884.2 ENST00000396928.1 |
SSX4B |
synovial sarcoma, X breakpoint 4B |
| chr7_+_99933730 | 0.06 |
ENST00000610247.1 |
PILRB |
paired immunoglobin-like type 2 receptor beta |
| chr1_-_48937821 | 0.06 |
ENST00000396199.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr17_+_73452545 | 0.06 |
ENST00000314256.7 |
KIAA0195 |
KIAA0195 |
| chr21_-_34185989 | 0.06 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr7_+_100209725 | 0.06 |
ENST00000223054.4 |
MOSPD3 |
motile sperm domain containing 3 |
| chr15_-_77712477 | 0.06 |
ENST00000560626.2 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
| chr12_-_120315074 | 0.06 |
ENST00000261833.7 ENST00000392521.2 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
| chr1_-_43855444 | 0.06 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr19_+_41770236 | 0.06 |
ENST00000392006.3 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_-_18266765 | 0.06 |
ENST00000354098.3 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr7_+_5919458 | 0.05 |
ENST00000416608.1 |
OCM |
oncomodulin |
| chr10_-_114206649 | 0.05 |
ENST00000369404.3 ENST00000369405.3 |
ZDHHC6 |
zinc finger, DHHC-type containing 6 |
| chrX_-_18690210 | 0.05 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr3_+_186353756 | 0.05 |
ENST00000431018.1 ENST00000450521.1 ENST00000539949.1 |
FETUB |
fetuin B |
| chrY_-_6740649 | 0.05 |
ENST00000383036.1 ENST00000383037.4 |
AMELY |
amelogenin, Y-linked |
| chr7_-_99277610 | 0.05 |
ENST00000343703.5 ENST00000222982.4 ENST00000439761.1 ENST00000339843.2 |
CYP3A5 |
cytochrome P450, family 3, subfamily A, polypeptide 5 |
| chr8_+_9953061 | 0.05 |
ENST00000522907.1 ENST00000528246.1 |
MSRA |
methionine sulfoxide reductase A |
| chr15_-_71055878 | 0.05 |
ENST00000322954.6 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr19_+_18699535 | 0.05 |
ENST00000358607.6 |
C19orf60 |
chromosome 19 open reading frame 60 |
| chr2_+_220143989 | 0.05 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr14_-_74551172 | 0.05 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr7_+_100209979 | 0.05 |
ENST00000493970.1 ENST00000379527.2 |
MOSPD3 |
motile sperm domain containing 3 |
| chr17_-_18266797 | 0.05 |
ENST00000316694.3 ENST00000539052.1 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr5_+_140220769 | 0.05 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr8_+_9953214 | 0.05 |
ENST00000382490.5 |
MSRA |
methionine sulfoxide reductase A |
| chr15_-_50411412 | 0.05 |
ENST00000284509.6 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
| chr11_+_76156045 | 0.05 |
ENST00000533988.1 ENST00000524490.1 ENST00000334736.3 ENST00000343878.3 ENST00000533972.1 |
C11orf30 |
chromosome 11 open reading frame 30 |
| chr6_+_64345698 | 0.05 |
ENST00000506783.1 ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3 |
PHD finger protein 3 |
| chr2_+_145780739 | 0.05 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr9_-_95166884 | 0.05 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr12_+_14927270 | 0.05 |
ENST00000544848.1 |
H2AFJ |
H2A histone family, member J |
| chr19_+_18699599 | 0.05 |
ENST00000450195.2 |
C19orf60 |
chromosome 19 open reading frame 60 |
| chr17_-_18950950 | 0.05 |
ENST00000284154.5 |
GRAP |
GRB2-related adaptor protein |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 3.0 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.2 | 0.7 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.2 | 1.2 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.2 | 0.6 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.6 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 0.3 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.5 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.1 | 1.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.2 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.1 | 1.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.9 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:0033383 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.1 | 0.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.2 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.2 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.0 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.4 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) response to antineoplastic agent(GO:0097327) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.3 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0034759 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:1903961 | positive regulation of anion transmembrane transport(GO:1903961) |
| 0.0 | 0.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 2.0 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.1 | GO:0072166 | posterior mesonephric tubule development(GO:0072166) negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.1 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.6 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 2.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.0 | GO:1990204 | NADPH oxidase complex(GO:0043020) oxidoreductase complex(GO:1990204) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 2.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.2 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.4 | 1.2 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 0.6 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 0.5 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.8 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 0.4 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.1 | 0.6 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.3 | GO:0047023 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.3 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.6 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 1.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.0 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.0 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |