Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
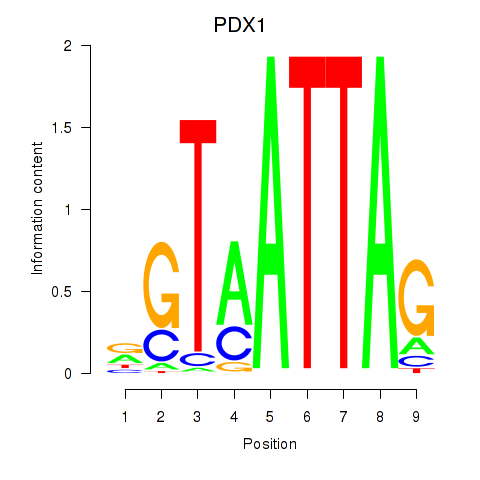
Results for PDX1
Z-value: 0.45
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | PDX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PDX1 | hg19_v2_chr13_+_28494130_28494168 | 0.17 | 5.3e-01 | Click! |
Activity profile of PDX1 motif
Sorted Z-values of PDX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PDX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_116660246 | 0.77 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr13_-_46716969 | 0.69 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_+_198608146 | 0.56 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chr6_-_32157947 | 0.54 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr3_-_38071122 | 0.49 |
ENST00000334661.4 |
PLCD1 |
phospholipase C, delta 1 |
| chr12_+_28410128 | 0.45 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr3_-_141747950 | 0.45 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_+_150920999 | 0.44 |
ENST00000367328.1 ENST00000367326.1 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr8_+_77593448 | 0.41 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr6_+_13272904 | 0.36 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr15_+_43985725 | 0.36 |
ENST00000413453.2 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr8_+_77593474 | 0.33 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr21_-_15918618 | 0.32 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr4_-_41884620 | 0.31 |
ENST00000504870.1 |
LINC00682 |
long intergenic non-protein coding RNA 682 |
| chr15_+_43886057 | 0.31 |
ENST00000441322.1 ENST00000413657.2 ENST00000453733.1 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr8_+_50824233 | 0.30 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr13_-_67802549 | 0.29 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr15_-_55563072 | 0.29 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_+_81110684 | 0.28 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr18_+_55888767 | 0.28 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr14_-_69261310 | 0.27 |
ENST00000336440.3 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chrX_-_110655306 | 0.25 |
ENST00000371993.2 |
DCX |
doublecortin |
| chr21_-_35899113 | 0.24 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr18_+_29027696 | 0.22 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr8_-_86290333 | 0.22 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr15_-_55562479 | 0.22 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr5_-_134735568 | 0.21 |
ENST00000510038.1 ENST00000304332.4 |
H2AFY |
H2A histone family, member Y |
| chr17_-_27418537 | 0.20 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr7_+_50348268 | 0.20 |
ENST00000438033.1 ENST00000439701.1 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr17_+_72427477 | 0.20 |
ENST00000342648.5 ENST00000481232.1 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
| chr14_+_22985251 | 0.20 |
ENST00000390510.1 |
TRAJ27 |
T cell receptor alpha joining 27 |
| chr3_+_111718173 | 0.20 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr1_+_28261533 | 0.20 |
ENST00000411604.1 ENST00000373888.4 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr14_+_72052983 | 0.19 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr3_+_69985734 | 0.19 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr12_-_16761007 | 0.19 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr4_-_155533787 | 0.18 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr16_-_29910853 | 0.18 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr1_+_62439037 | 0.17 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr12_-_86650045 | 0.17 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr8_-_42234745 | 0.17 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chrX_-_20236970 | 0.17 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr12_-_86650077 | 0.17 |
ENST00000552808.2 ENST00000547225.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr1_-_190446759 | 0.17 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr4_-_116034979 | 0.17 |
ENST00000264363.2 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
| chr12_-_52967600 | 0.17 |
ENST00000549343.1 ENST00000305620.2 |
KRT74 |
keratin 74 |
| chr11_-_8290263 | 0.16 |
ENST00000428101.2 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
| chr3_+_69985792 | 0.15 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr20_+_52105495 | 0.15 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr11_-_115630900 | 0.15 |
ENST00000537070.1 ENST00000499809.1 ENST00000514294.2 ENST00000535683.1 |
LINC00900 |
long intergenic non-protein coding RNA 900 |
| chr2_-_136678123 | 0.15 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr4_+_144354644 | 0.14 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr7_-_25268104 | 0.14 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr1_-_1709845 | 0.14 |
ENST00000341426.5 ENST00000344463.4 |
NADK |
NAD kinase |
| chr3_+_111718036 | 0.14 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr3_-_57233966 | 0.14 |
ENST00000473921.1 ENST00000295934.3 |
HESX1 |
HESX homeobox 1 |
| chr1_+_199996702 | 0.13 |
ENST00000367362.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr6_+_34204642 | 0.13 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr2_-_166930131 | 0.13 |
ENST00000303395.4 ENST00000409050.1 ENST00000423058.2 ENST00000375405.3 |
SCN1A |
sodium channel, voltage-gated, type I, alpha subunit |
| chr4_-_103749205 | 0.13 |
ENST00000508249.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr12_+_41831485 | 0.13 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr4_-_89978299 | 0.13 |
ENST00000511976.1 ENST00000509094.1 ENST00000264344.5 ENST00000515600.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr14_-_36988882 | 0.12 |
ENST00000498187.2 |
NKX2-1 |
NK2 homeobox 1 |
| chr1_-_38019878 | 0.12 |
ENST00000296215.6 |
SNIP1 |
Smad nuclear interacting protein 1 |
| chr16_-_30457048 | 0.12 |
ENST00000500504.2 ENST00000542752.1 |
SEPHS2 |
selenophosphate synthetase 2 |
| chr13_-_41593425 | 0.12 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr1_+_68150744 | 0.12 |
ENST00000370986.4 ENST00000370985.3 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
| chr9_-_124990680 | 0.11 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chr3_-_112127981 | 0.11 |
ENST00000486726.2 |
RP11-231E6.1 |
RP11-231E6.1 |
| chr1_+_101003687 | 0.11 |
ENST00000315033.4 |
GPR88 |
G protein-coupled receptor 88 |
| chr13_-_28545276 | 0.11 |
ENST00000381020.7 |
CDX2 |
caudal type homeobox 2 |
| chr15_+_93443419 | 0.11 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr1_-_1710229 | 0.10 |
ENST00000341991.3 |
NADK |
NAD kinase |
| chr20_+_62795827 | 0.10 |
ENST00000328439.1 ENST00000536311.1 |
MYT1 |
myelin transcription factor 1 |
| chr4_-_66536196 | 0.10 |
ENST00000511294.1 |
EPHA5 |
EPH receptor A5 |
| chr2_-_190927447 | 0.10 |
ENST00000260950.4 |
MSTN |
myostatin |
| chr18_-_33709268 | 0.10 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr12_+_81101277 | 0.10 |
ENST00000228641.3 |
MYF6 |
myogenic factor 6 (herculin) |
| chr2_-_145277569 | 0.10 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr1_-_212965104 | 0.10 |
ENST00000422588.2 ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
| chr6_-_109702885 | 0.10 |
ENST00000504373.1 |
CD164 |
CD164 molecule, sialomucin |
| chr4_-_123377880 | 0.10 |
ENST00000226730.4 |
IL2 |
interleukin 2 |
| chr2_+_45168875 | 0.09 |
ENST00000260653.3 |
SIX3 |
SIX homeobox 3 |
| chr15_-_65903407 | 0.09 |
ENST00000395644.4 ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9 |
von Willebrand factor A domain containing 9 |
| chr4_-_66536057 | 0.09 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr4_-_103749179 | 0.09 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_108541545 | 0.09 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr1_+_50574585 | 0.09 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr16_+_6533729 | 0.09 |
ENST00000551752.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chrX_-_130423200 | 0.09 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr3_+_108541608 | 0.08 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr12_-_52828147 | 0.08 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr10_+_119301928 | 0.08 |
ENST00000553456.3 |
EMX2 |
empty spiracles homeobox 2 |
| chr14_-_61116168 | 0.08 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr20_+_42984330 | 0.08 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr3_+_111717600 | 0.08 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr5_-_140998616 | 0.08 |
ENST00000389054.3 ENST00000398562.2 ENST00000389057.5 ENST00000398566.3 ENST00000398557.4 ENST00000253811.6 |
DIAPH1 |
diaphanous-related formin 1 |
| chr5_-_140998481 | 0.08 |
ENST00000518047.1 |
DIAPH1 |
diaphanous-related formin 1 |
| chr3_-_101039402 | 0.08 |
ENST00000193391.7 |
IMPG2 |
interphotoreceptor matrix proteoglycan 2 |
| chr3_+_111717511 | 0.07 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr14_+_100485712 | 0.07 |
ENST00000544450.2 |
EVL |
Enah/Vasp-like |
| chr5_-_141249154 | 0.07 |
ENST00000357517.5 ENST00000536585.1 |
PCDH1 |
protocadherin 1 |
| chr11_-_129062093 | 0.07 |
ENST00000310343.9 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr17_-_40337470 | 0.07 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr5_-_142780280 | 0.07 |
ENST00000424646.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr20_+_30697298 | 0.07 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr11_+_125034586 | 0.07 |
ENST00000298282.9 |
PKNOX2 |
PBX/knotted 1 homeobox 2 |
| chr1_-_10532531 | 0.07 |
ENST00000377036.2 ENST00000377038.3 |
DFFA |
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr6_+_45296391 | 0.07 |
ENST00000371436.6 ENST00000576263.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr18_+_28898052 | 0.07 |
ENST00000257192.4 |
DSG1 |
desmoglein 1 |
| chr19_+_11071546 | 0.07 |
ENST00000358026.2 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr4_+_151503077 | 0.06 |
ENST00000317605.4 |
MAB21L2 |
mab-21-like 2 (C. elegans) |
| chr3_+_115342349 | 0.06 |
ENST00000393780.3 |
GAP43 |
growth associated protein 43 |
| chr15_-_34875771 | 0.06 |
ENST00000267731.7 |
GOLGA8B |
golgin A8 family, member B |
| chr12_-_53171128 | 0.06 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chr7_+_129015484 | 0.06 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr18_+_3447572 | 0.06 |
ENST00000548489.2 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr4_-_39979576 | 0.06 |
ENST00000303538.8 ENST00000503396.1 |
PDS5A |
PDS5, regulator of cohesion maintenance, homolog A (S. cerevisiae) |
| chr3_-_167191814 | 0.06 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr2_+_196313239 | 0.06 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr2_+_48541776 | 0.06 |
ENST00000413569.1 ENST00000340553.3 |
FOXN2 |
forkhead box N2 |
| chr5_-_134871639 | 0.06 |
ENST00000314744.4 |
NEUROG1 |
neurogenin 1 |
| chr15_+_62853562 | 0.06 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr1_-_13002348 | 0.06 |
ENST00000355096.2 |
PRAMEF6 |
PRAME family member 6 |
| chr15_-_55562582 | 0.06 |
ENST00000396307.2 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr19_+_641178 | 0.06 |
ENST00000166133.3 |
FGF22 |
fibroblast growth factor 22 |
| chr11_+_65554493 | 0.05 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr17_-_6983594 | 0.05 |
ENST00000571664.1 ENST00000254868.4 |
CLEC10A |
C-type lectin domain family 10, member A |
| chr11_-_8285405 | 0.05 |
ENST00000335790.3 ENST00000534484.1 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
| chr6_+_29079668 | 0.05 |
ENST00000377169.1 |
OR2J3 |
olfactory receptor, family 2, subfamily J, member 3 |
| chr12_+_49621658 | 0.05 |
ENST00000541364.1 |
TUBA1C |
tubulin, alpha 1c |
| chr7_-_99717463 | 0.05 |
ENST00000437822.2 |
TAF6 |
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr16_+_6533380 | 0.05 |
ENST00000552089.1 |
RBFOX1 |
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr15_+_67841330 | 0.05 |
ENST00000354498.5 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
| chr2_-_61697862 | 0.05 |
ENST00000398571.2 |
USP34 |
ubiquitin specific peptidase 34 |
| chr5_+_174151536 | 0.05 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr12_+_50144381 | 0.05 |
ENST00000552370.1 |
TMBIM6 |
transmembrane BAX inhibitor motif containing 6 |
| chr13_-_99667960 | 0.05 |
ENST00000448493.2 |
DOCK9 |
dedicator of cytokinesis 9 |
| chr11_-_115375107 | 0.05 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr20_+_43029911 | 0.05 |
ENST00000443598.2 ENST00000316099.4 ENST00000415691.2 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr4_-_53525406 | 0.05 |
ENST00000451218.2 ENST00000441222.3 |
USP46 |
ubiquitin specific peptidase 46 |
| chr8_-_133123406 | 0.05 |
ENST00000434736.2 |
HHLA1 |
HERV-H LTR-associating 1 |
| chr1_-_2458026 | 0.05 |
ENST00000435556.3 ENST00000378466.3 |
PANK4 |
pantothenate kinase 4 |
| chr7_-_44229022 | 0.05 |
ENST00000403799.3 |
GCK |
glucokinase (hexokinase 4) |
| chr17_-_46690839 | 0.04 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr4_-_103748880 | 0.04 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr6_-_25042231 | 0.04 |
ENST00000510784.2 |
FAM65B |
family with sequence similarity 65, member B |
| chr18_+_59000815 | 0.04 |
ENST00000262717.4 |
CDH20 |
cadherin 20, type 2 |
| chr12_-_56106060 | 0.04 |
ENST00000452168.2 |
ITGA7 |
integrin, alpha 7 |
| chr9_-_14722715 | 0.04 |
ENST00000380911.3 |
CER1 |
cerberus 1, DAN family BMP antagonist |
| chr7_+_100860949 | 0.03 |
ENST00000305105.2 |
ZNHIT1 |
zinc finger, HIT-type containing 1 |
| chr17_-_6983550 | 0.03 |
ENST00000576617.1 ENST00000416562.2 |
CLEC10A |
C-type lectin domain family 10, member A |
| chr6_-_38670897 | 0.03 |
ENST00000373365.4 |
GLO1 |
glyoxalase I |
| chr15_+_48498480 | 0.03 |
ENST00000380993.3 ENST00000396577.3 |
SLC12A1 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 1 |
| chr2_+_166152283 | 0.03 |
ENST00000375427.2 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr20_+_60174827 | 0.03 |
ENST00000543233.1 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
| chr15_+_75080883 | 0.03 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr4_-_41750922 | 0.03 |
ENST00000226382.2 |
PHOX2B |
paired-like homeobox 2b |
| chr1_+_159272111 | 0.03 |
ENST00000368114.1 |
FCER1A |
Fc fragment of IgE, high affinity I, receptor for; alpha polypeptide |
| chr12_-_51611477 | 0.02 |
ENST00000389243.4 |
POU6F1 |
POU class 6 homeobox 1 |
| chr15_+_34261089 | 0.02 |
ENST00000383263.5 |
CHRM5 |
cholinergic receptor, muscarinic 5 |
| chr5_+_169532896 | 0.02 |
ENST00000306268.6 ENST00000449804.2 |
FOXI1 |
forkhead box I1 |
| chr6_-_100912785 | 0.02 |
ENST00000369208.3 |
SIM1 |
single-minded family bHLH transcription factor 1 |
| chr3_+_35722487 | 0.02 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chrX_-_13835147 | 0.02 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr6_-_33160231 | 0.02 |
ENST00000395194.1 ENST00000457788.1 ENST00000341947.2 ENST00000357486.1 ENST00000374714.1 ENST00000374713.1 ENST00000395197.1 ENST00000374712.1 ENST00000361917.1 ENST00000374708.4 |
COL11A2 |
collagen, type XI, alpha 2 |
| chrX_-_100129128 | 0.02 |
ENST00000372960.4 ENST00000372964.1 ENST00000217885.5 |
NOX1 |
NADPH oxidase 1 |
| chr17_-_9929581 | 0.02 |
ENST00000437099.2 ENST00000396115.2 |
GAS7 |
growth arrest-specific 7 |
| chr15_+_58430368 | 0.02 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr7_-_151217001 | 0.02 |
ENST00000262187.5 |
RHEB |
Ras homolog enriched in brain |
| chr6_-_87804815 | 0.01 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr15_+_58430567 | 0.01 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr1_+_45241109 | 0.01 |
ENST00000396651.3 |
RPS8 |
ribosomal protein S8 |
| chr11_-_36619771 | 0.01 |
ENST00000311485.3 ENST00000527033.1 ENST00000532616.1 |
RAG2 |
recombination activating gene 2 |
| chr5_-_9630463 | 0.01 |
ENST00000382492.2 |
TAS2R1 |
taste receptor, type 2, member 1 |
| chr17_+_60536002 | 0.01 |
ENST00000582809.1 |
TLK2 |
tousled-like kinase 2 |
| chr3_-_108248169 | 0.01 |
ENST00000273353.3 |
MYH15 |
myosin, heavy chain 15 |
| chr15_-_31393910 | 0.01 |
ENST00000397795.2 ENST00000256552.6 ENST00000559179.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chrX_+_144908928 | 0.01 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr4_-_119759795 | 0.01 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr12_+_15699286 | 0.01 |
ENST00000442921.2 ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr15_-_72563585 | 0.01 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr10_-_1779663 | 0.01 |
ENST00000381312.1 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr7_-_14028488 | 0.01 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr2_+_109204743 | 0.01 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr4_-_103749313 | 0.01 |
ENST00000394803.5 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr10_+_114710425 | 0.00 |
ENST00000352065.5 ENST00000369395.1 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr10_-_48416849 | 0.00 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr3_+_149191723 | 0.00 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr6_-_111136513 | 0.00 |
ENST00000368911.3 |
CDK19 |
cyclin-dependent kinase 19 |
| chr11_-_102496063 | 0.00 |
ENST00000260228.2 |
MMP20 |
matrix metallopeptidase 20 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.6 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.1 | 0.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 0.3 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:1901837 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.1 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.5 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.4 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.2 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) myotome development(GO:0061055) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) negative regulation of skeletal muscle cell proliferation(GO:0014859) regulation of skeletal muscle tissue growth(GO:0048631) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.5 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) regulation of catagen(GO:0051794) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) activation of meiosis(GO:0090427) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.0 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.5 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.2 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.0 | 0.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.5 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |