Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for PGR
Z-value: 1.00
Transcription factors associated with PGR
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PGR
|
ENSG00000082175.10 | PGR |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PGR | hg19_v2_chr11_-_101000445_101000465, hg19_v2_chr11_-_100999775_100999801 | -0.14 | 6.0e-01 | Click! |
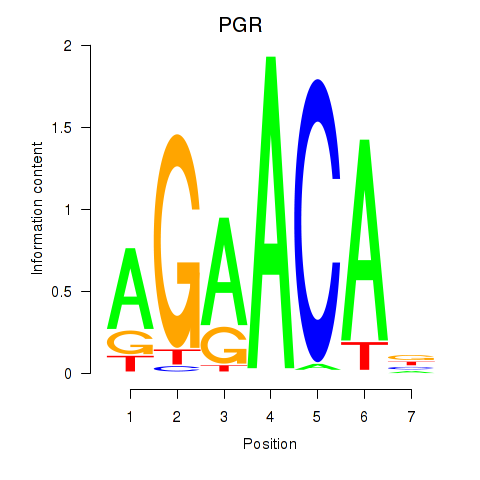
Activity profile of PGR motif
Sorted Z-values of PGR motif
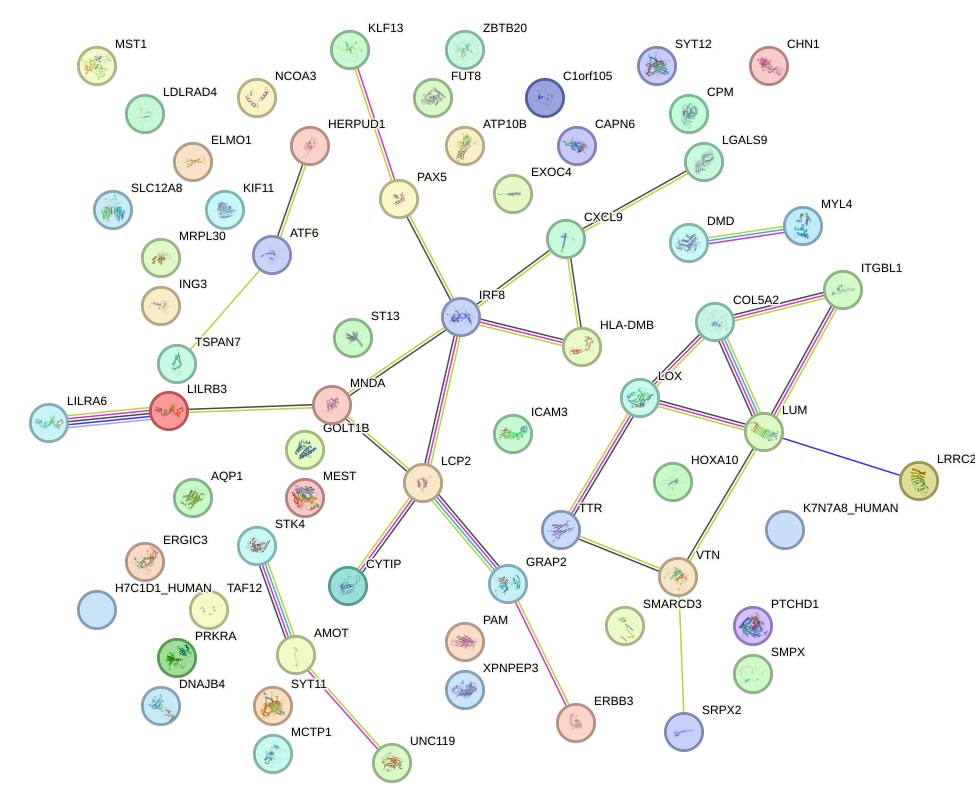
Network of associatons between targets according to the STRING database.
First level regulatory network of PGR
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_102104980 | 2.77 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_-_114343768 | 2.69 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr13_+_102104952 | 2.50 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr3_-_114035026 | 2.18 |
ENST00000570269.1 |
RP11-553L6.5 |
RP11-553L6.5 |
| chr1_-_201390846 | 2.14 |
ENST00000367312.1 ENST00000555340.2 ENST00000361379.4 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr7_+_130126012 | 2.00 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr12_-_91505608 | 1.75 |
ENST00000266718.4 |
LUM |
lumican |
| chr3_-_114343039 | 1.72 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr1_-_201391149 | 1.72 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr6_-_32908792 | 1.40 |
ENST00000418107.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr14_-_106692191 | 1.34 |
ENST00000390607.2 |
IGHV3-21 |
immunoglobulin heavy variable 3-21 |
| chr7_+_130126165 | 1.27 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr2_-_175711133 | 1.22 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr2_-_158300556 | 1.21 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chrX_-_21776281 | 1.18 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr11_-_73693875 | 1.17 |
ENST00000536983.1 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr16_+_56970567 | 1.13 |
ENST00000563911.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr16_+_32077386 | 1.10 |
ENST00000354689.6 |
IGHV3OR16-9 |
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr7_-_37026108 | 1.05 |
ENST00000396045.3 |
ELMO1 |
engulfment and cell motility 1 |
| chr2_-_179672142 | 0.97 |
ENST00000342992.6 ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN |
titin |
| chr3_-_124839648 | 0.95 |
ENST00000430155.2 |
SLC12A8 |
solute carrier family 12, member 8 |
| chr1_+_158815588 | 0.92 |
ENST00000438394.1 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr1_+_111770278 | 0.90 |
ENST00000369748.4 |
CHI3L2 |
chitinase 3-like 2 |
| chr1_+_111770232 | 0.89 |
ENST00000369744.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr2_-_190044480 | 0.86 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr4_-_71532339 | 0.84 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr19_-_10446449 | 0.79 |
ENST00000592439.1 |
ICAM3 |
intercellular adhesion molecule 3 |
| chr5_-_121413974 | 0.78 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chrX_+_38420623 | 0.73 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr16_+_85942594 | 0.71 |
ENST00000566369.1 |
IRF8 |
interferon regulatory factor 8 |
| chr14_-_107219365 | 0.68 |
ENST00000424969.2 |
IGHV3-74 |
immunoglobulin heavy variable 3-74 |
| chrX_+_99899180 | 0.67 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr14_-_61124977 | 0.67 |
ENST00000554986.1 |
SIX1 |
SIX homeobox 1 |
| chr16_+_33605231 | 0.65 |
ENST00000570121.2 |
IGHV3OR16-12 |
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr13_-_46716969 | 0.64 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr9_-_37034028 | 0.62 |
ENST00000520281.1 ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5 |
paired box 5 |
| chr9_-_15510989 | 0.62 |
ENST00000380715.1 ENST00000380716.4 ENST00000380738.4 ENST00000380733.4 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr1_+_155829286 | 0.61 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr7_-_150946015 | 0.61 |
ENST00000262188.8 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr7_+_73442487 | 0.60 |
ENST00000380575.4 ENST00000380584.4 ENST00000458204.1 ENST00000357036.5 ENST00000417091.1 ENST00000429192.1 ENST00000442310.1 ENST00000380553.4 ENST00000380576.5 ENST00000428787.1 ENST00000320399.6 |
ELN |
elastin |
| chr7_-_27213893 | 0.60 |
ENST00000283921.4 |
HOXA10 |
homeobox A10 |
| chr7_+_73442422 | 0.58 |
ENST00000358929.4 ENST00000431562.1 ENST00000320492.7 ENST00000438906.1 |
ELN |
elastin |
| chrX_-_100641155 | 0.53 |
ENST00000372880.1 ENST00000308731.7 |
BTK |
Bruton agammaglobulinemia tyrosine kinase |
| chr17_+_45286387 | 0.53 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr1_+_78470530 | 0.52 |
ENST00000370763.5 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr6_+_39760129 | 0.51 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr14_+_65878565 | 0.51 |
ENST00000556518.1 ENST00000557164.1 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
| chr5_+_102201509 | 0.50 |
ENST00000348126.2 ENST00000379787.4 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr2_+_90153696 | 0.47 |
ENST00000417279.2 |
IGKV3D-15 |
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr2_-_179315786 | 0.44 |
ENST00000457633.1 ENST00000438687.3 ENST00000325748.4 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr17_+_45286706 | 0.42 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr11_+_20044600 | 0.42 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr6_-_32908765 | 0.41 |
ENST00000416244.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr12_+_56473628 | 0.41 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr7_+_73442457 | 0.40 |
ENST00000438880.1 ENST00000414324.1 ENST00000380562.4 |
ELN |
elastin |
| chrX_+_54834004 | 0.39 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr13_+_43148281 | 0.38 |
ENST00000239849.6 ENST00000398795.2 ENST00000544862.1 |
TNFSF11 |
tumor necrosis factor (ligand) superfamily, member 11 |
| chr20_+_46130619 | 0.38 |
ENST00000372004.3 |
NCOA3 |
nuclear receptor coactivator 3 |
| chr1_+_172422026 | 0.37 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr1_-_154164534 | 0.36 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr8_+_22446763 | 0.36 |
ENST00000450780.2 ENST00000430850.2 ENST00000447849.1 |
AC037459.4 |
Uncharacterized protein |
| chr9_+_116298778 | 0.36 |
ENST00000462143.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr20_+_34129770 | 0.36 |
ENST00000348547.2 ENST00000357394.4 ENST00000447986.1 ENST00000279052.6 ENST00000416206.1 ENST00000411577.1 ENST00000413587.1 |
ERGIC3 |
ERGIC and golgi 3 |
| chr9_+_116225999 | 0.36 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr17_+_25958174 | 0.36 |
ENST00000313648.6 ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9 |
lectin, galactoside-binding, soluble, 9 |
| chr11_-_615570 | 0.34 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chr5_-_94417339 | 0.33 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr12_-_69326940 | 0.33 |
ENST00000549781.1 ENST00000548262.1 ENST00000551568.1 ENST00000548954.1 |
CPM |
carboxypeptidase M |
| chr4_-_105416039 | 0.33 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr14_-_23876801 | 0.33 |
ENST00000356287.3 |
MYH6 |
myosin, heavy chain 6, cardiac muscle, alpha |
| chr22_+_40297079 | 0.32 |
ENST00000344138.4 ENST00000543252.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr17_+_34136459 | 0.32 |
ENST00000588240.1 ENST00000590273.1 ENST00000588441.1 ENST00000587272.1 ENST00000592237.1 ENST00000311979.3 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr17_-_26876350 | 0.31 |
ENST00000470125.1 |
UNC119 |
unc-119 homolog (C. elegans) |
| chr5_+_102201722 | 0.31 |
ENST00000274392.9 ENST00000455264.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chr22_+_40297105 | 0.30 |
ENST00000540310.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr1_-_115292591 | 0.30 |
ENST00000438362.2 |
CSDE1 |
cold shock domain containing E1, RNA-binding |
| chrX_-_112084043 | 0.30 |
ENST00000304758.1 |
AMOT |
angiomotin |
| chr18_+_13611763 | 0.30 |
ENST00000585931.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr5_+_102201430 | 0.29 |
ENST00000438793.3 ENST00000346918.2 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
| chrX_-_32173579 | 0.29 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr9_+_2159850 | 0.29 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr10_+_94352956 | 0.28 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr15_+_31658349 | 0.28 |
ENST00000558844.1 |
KLF13 |
Kruppel-like factor 13 |
| chr9_+_82188077 | 0.27 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr17_-_26879567 | 0.27 |
ENST00000581945.1 ENST00000444148.1 ENST00000301032.4 ENST00000335765.4 |
UNC119 |
unc-119 homolog (C. elegans) |
| chr19_+_35773242 | 0.27 |
ENST00000222304.3 |
HAMP |
hepcidin antimicrobial peptide |
| chr1_+_161736072 | 0.26 |
ENST00000367942.3 |
ATF6 |
activating transcription factor 6 |
| chr2_-_179315453 | 0.26 |
ENST00000432031.2 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr7_+_120590803 | 0.26 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr2_-_179315490 | 0.26 |
ENST00000487082.1 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
| chr7_+_73442102 | 0.26 |
ENST00000445912.1 ENST00000252034.7 |
ELN |
elastin |
| chrX_+_46940254 | 0.25 |
ENST00000336169.3 |
RGN |
regucalcin |
| chr22_+_41253080 | 0.25 |
ENST00000541156.1 ENST00000414396.1 ENST00000357137.4 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr14_+_24641062 | 0.24 |
ENST00000311457.3 ENST00000557806.1 ENST00000559919.1 |
REC8 |
REC8 meiotic recombination protein |
| chr5_-_160279207 | 0.24 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr11_+_123396528 | 0.24 |
ENST00000322282.7 ENST00000529750.1 |
GRAMD1B |
GRAM domain containing 1B |
| chr10_+_73156664 | 0.23 |
ENST00000398809.4 ENST00000398842.3 ENST00000461841.3 ENST00000299366.7 |
CDH23 |
cadherin-related 23 |
| chr4_-_76928641 | 0.23 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr2_+_99797542 | 0.23 |
ENST00000338148.3 ENST00000512183.2 |
MRPL30 C2orf15 |
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chr18_+_29171689 | 0.23 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr12_+_21654714 | 0.23 |
ENST00000542038.1 ENST00000540141.1 ENST00000229314.5 |
GOLT1B |
golgi transport 1B |
| chr7_+_30960915 | 0.23 |
ENST00000441328.2 ENST00000409899.1 ENST00000409611.1 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chr2_+_201173667 | 0.22 |
ENST00000409755.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr14_-_23877474 | 0.21 |
ENST00000405093.3 |
MYH6 |
myosin, heavy chain 6, cardiac muscle, alpha |
| chr10_-_72142345 | 0.21 |
ENST00000373224.1 ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20 |
leucine rich repeat containing 20 |
| chr17_-_26694979 | 0.20 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr5_-_169694286 | 0.20 |
ENST00000521416.1 ENST00000520344.1 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chrX_-_110513703 | 0.20 |
ENST00000324068.1 |
CAPN6 |
calpain 6 |
| chr22_-_41252962 | 0.20 |
ENST00000216218.3 |
ST13 |
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr19_-_54726850 | 0.19 |
ENST00000245620.9 ENST00000346401.6 ENST00000424807.1 ENST00000445347.1 |
LILRB3 |
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
| chr3_-_52001448 | 0.18 |
ENST00000461554.1 ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chrX_+_23352133 | 0.18 |
ENST00000379361.4 |
PTCHD1 |
patched domain containing 1 |
| chr7_+_22766766 | 0.18 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr2_+_191208196 | 0.18 |
ENST00000392329.2 ENST00000322522.4 ENST00000430311.1 ENST00000541441.1 |
INPP1 |
inositol polyphosphate-1-phosphatase |
| chr3_-_49722523 | 0.17 |
ENST00000448220.1 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr19_+_49377575 | 0.17 |
ENST00000600406.1 |
PPP1R15A |
protein phosphatase 1, regulatory subunit 15A |
| chrX_+_54834159 | 0.16 |
ENST00000375053.2 ENST00000347546.4 ENST00000375062.4 |
MAGED2 |
melanoma antigen family D, 2 |
| chr5_+_140797296 | 0.16 |
ENST00000398594.2 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
| chr2_+_191334212 | 0.16 |
ENST00000444317.1 ENST00000535751.1 |
MFSD6 |
major facilitator superfamily domain containing 6 |
| chr16_+_33006369 | 0.16 |
ENST00000425181.3 |
IGHV3OR16-10 |
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr19_+_16771936 | 0.16 |
ENST00000187762.2 ENST00000599479.1 |
TMEM38A |
transmembrane protein 38A |
| chr17_+_73997796 | 0.15 |
ENST00000586261.1 |
CDK3 |
cyclin-dependent kinase 3 |
| chr19_-_8642289 | 0.15 |
ENST00000596675.1 ENST00000338257.8 |
MYO1F |
myosin IF |
| chr2_+_11674213 | 0.15 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr7_-_135433460 | 0.15 |
ENST00000415751.1 |
FAM180A |
family with sequence similarity 180, member A |
| chr14_-_64194745 | 0.15 |
ENST00000247225.6 |
SGPP1 |
sphingosine-1-phosphate phosphatase 1 |
| chr3_-_195270162 | 0.14 |
ENST00000438848.1 ENST00000328432.3 |
PPP1R2 |
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr2_-_89266286 | 0.13 |
ENST00000464162.1 |
IGKV1-6 |
immunoglobulin kappa variable 1-6 |
| chr5_+_138629417 | 0.13 |
ENST00000510056.1 ENST00000511249.1 ENST00000503811.1 ENST00000511378.1 |
MATR3 |
matrin 3 |
| chr20_+_35090150 | 0.13 |
ENST00000340491.4 |
DLGAP4 |
discs, large (Drosophila) homolog-associated protein 4 |
| chr15_+_31619013 | 0.13 |
ENST00000307145.3 |
KLF13 |
Kruppel-like factor 13 |
| chr5_+_138629337 | 0.13 |
ENST00000394805.3 ENST00000512876.1 ENST00000513678.1 |
MATR3 |
matrin 3 |
| chr12_+_57849048 | 0.12 |
ENST00000266646.2 |
INHBE |
inhibin, beta E |
| chr17_+_59477233 | 0.12 |
ENST00000240328.3 |
TBX2 |
T-box 2 |
| chr6_-_154831779 | 0.12 |
ENST00000607772.1 |
CNKSR3 |
CNKSR family member 3 |
| chr22_-_30162924 | 0.12 |
ENST00000344318.3 ENST00000397781.3 |
ZMAT5 |
zinc finger, matrin-type 5 |
| chr19_-_6333614 | 0.12 |
ENST00000301452.4 |
ACER1 |
alkaline ceramidase 1 |
| chr3_+_29323043 | 0.12 |
ENST00000452462.1 ENST00000456853.1 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
| chr3_-_149470229 | 0.12 |
ENST00000473414.1 |
COMMD2 |
COMM domain containing 2 |
| chr4_-_146019693 | 0.11 |
ENST00000514390.1 |
ANAPC10 |
anaphase promoting complex subunit 10 |
| chr11_+_57105991 | 0.11 |
ENST00000263314.2 |
P2RX3 |
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr18_-_44336998 | 0.11 |
ENST00000315087.7 |
ST8SIA5 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr6_-_43021437 | 0.11 |
ENST00000265348.3 |
CUL7 |
cullin 7 |
| chr3_+_137906109 | 0.10 |
ENST00000481646.1 ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr3_+_137906353 | 0.10 |
ENST00000461822.1 ENST00000485396.1 ENST00000471453.1 ENST00000470821.1 ENST00000471709.1 ENST00000538260.1 ENST00000393058.3 ENST00000463485.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr18_+_6729725 | 0.10 |
ENST00000400091.2 ENST00000583410.1 ENST00000584387.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr1_-_32210275 | 0.10 |
ENST00000440175.2 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
| chrX_-_114953669 | 0.10 |
ENST00000449327.1 |
RP1-241P17.4 |
Uncharacterized protein |
| chr1_-_145039949 | 0.10 |
ENST00000313382.9 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr17_-_4464081 | 0.10 |
ENST00000574154.1 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr7_-_76829125 | 0.09 |
ENST00000248598.5 |
FGL2 |
fibrinogen-like 2 |
| chr1_+_155146318 | 0.09 |
ENST00000368385.4 ENST00000545012.1 ENST00000392451.2 ENST00000368383.3 ENST00000368382.1 ENST00000334634.4 |
TRIM46 |
tripartite motif containing 46 |
| chr1_-_167906277 | 0.09 |
ENST00000271373.4 |
MPC2 |
mitochondrial pyruvate carrier 2 |
| chr1_-_110933663 | 0.09 |
ENST00000369781.4 ENST00000541986.1 ENST00000369779.4 |
SLC16A4 |
solute carrier family 16, member 4 |
| chrX_+_99839799 | 0.08 |
ENST00000373031.4 |
TNMD |
tenomodulin |
| chr19_-_40440533 | 0.08 |
ENST00000221347.6 |
FCGBP |
Fc fragment of IgG binding protein |
| chr3_+_137906154 | 0.08 |
ENST00000466749.1 ENST00000358441.2 ENST00000489213.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr17_-_40540586 | 0.08 |
ENST00000264657.5 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr19_+_41768561 | 0.08 |
ENST00000599719.1 ENST00000601309.1 |
HNRNPUL1 |
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr19_+_52264449 | 0.08 |
ENST00000599326.1 ENST00000598953.1 |
FPR2 |
formyl peptide receptor 2 |
| chr16_-_67517716 | 0.08 |
ENST00000290953.2 |
AGRP |
agouti related protein homolog (mouse) |
| chr11_-_65625330 | 0.08 |
ENST00000531407.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr2_+_73144604 | 0.07 |
ENST00000258106.6 |
EMX1 |
empty spiracles homeobox 1 |
| chr5_-_148442584 | 0.07 |
ENST00000394358.2 ENST00000512049.1 |
SH3TC2 |
SH3 domain and tetratricopeptide repeats 2 |
| chr6_-_76203345 | 0.07 |
ENST00000393004.2 |
FILIP1 |
filamin A interacting protein 1 |
| chr6_+_167412835 | 0.07 |
ENST00000349556.4 |
FGFR1OP |
FGFR1 oncogene partner |
| chr6_+_31674639 | 0.07 |
ENST00000556581.1 ENST00000375832.4 ENST00000503322.1 |
LY6G6F MEGT1 |
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr12_+_125478241 | 0.07 |
ENST00000341446.8 |
BRI3BP |
BRI3 binding protein |
| chr7_-_5821314 | 0.07 |
ENST00000425013.2 ENST00000389902.3 |
RNF216 |
ring finger protein 216 |
| chr15_+_67841330 | 0.07 |
ENST00000354498.5 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
| chr17_-_26697304 | 0.07 |
ENST00000536498.1 |
VTN |
vitronectin |
| chr15_-_74726283 | 0.06 |
ENST00000543145.2 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr21_+_34398153 | 0.06 |
ENST00000382357.3 ENST00000430860.1 ENST00000333337.3 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
| chr10_-_28571015 | 0.06 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr7_+_105603657 | 0.06 |
ENST00000542731.1 ENST00000343407.5 |
CDHR3 |
cadherin-related family member 3 |
| chr12_+_122242597 | 0.06 |
ENST00000267197.5 |
SETD1B |
SET domain containing 1B |
| chr13_+_32838801 | 0.06 |
ENST00000542859.1 |
FRY |
furry homolog (Drosophila) |
| chr18_+_43304092 | 0.06 |
ENST00000321925.4 ENST00000587601.1 |
SLC14A1 |
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr5_-_115872142 | 0.06 |
ENST00000510263.1 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_+_110527308 | 0.05 |
ENST00000369799.5 |
AHCYL1 |
adenosylhomocysteinase-like 1 |
| chr13_-_52027134 | 0.05 |
ENST00000311234.4 ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6 |
integrator complex subunit 6 |
| chr10_+_124913930 | 0.05 |
ENST00000368858.5 |
BUB3 |
BUB3 mitotic checkpoint protein |
| chr4_-_71705060 | 0.05 |
ENST00000514161.1 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr18_+_6729698 | 0.05 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr2_-_219925189 | 0.05 |
ENST00000295731.6 |
IHH |
indian hedgehog |
| chr17_-_4463856 | 0.05 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr1_-_179112189 | 0.05 |
ENST00000512653.1 ENST00000344730.3 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr17_+_41150793 | 0.05 |
ENST00000586277.1 |
RPL27 |
ribosomal protein L27 |
| chr10_-_48416849 | 0.04 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr22_+_42017987 | 0.04 |
ENST00000405506.1 |
XRCC6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr4_-_139163491 | 0.04 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr8_-_42623747 | 0.04 |
ENST00000534622.1 |
CHRNA6 |
cholinergic receptor, nicotinic, alpha 6 (neuronal) |
| chr19_-_15443318 | 0.04 |
ENST00000360016.5 |
BRD4 |
bromodomain containing 4 |
| chr19_+_19431490 | 0.04 |
ENST00000392313.6 ENST00000262815.8 ENST00000609122.1 |
MAU2 |
MAU2 sister chromatid cohesion factor |
| chr2_-_165697920 | 0.04 |
ENST00000342193.4 ENST00000375458.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr1_+_16370271 | 0.04 |
ENST00000375679.4 |
CLCNKB |
chloride channel, voltage-sensitive Kb |
| chr18_+_32173276 | 0.04 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr1_-_89357179 | 0.04 |
ENST00000448623.1 ENST00000418217.1 ENST00000370500.5 |
GTF2B |
general transcription factor IIB |
| chr16_+_86600857 | 0.03 |
ENST00000320354.4 |
FOXC2 |
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
| chr15_-_76005170 | 0.03 |
ENST00000308508.5 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.3 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.8 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 0.9 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.2 | 0.5 | GO:0008424 | glycoprotein 6-alpha-L-fucosyltransferase activity(GO:0008424) alpha-(1->6)-fucosyltransferase activity(GO:0046921) |
| 0.2 | 1.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 1.8 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.1 | 1.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.0 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.1 | 0.5 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.1 | 1.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 3.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.8 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 0.8 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.6 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.1 | 0.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.2 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 4.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 5.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.6 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.3 | 1.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 1.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.3 | 1.0 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.2 | 3.9 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.2 | 0.7 | GO:0061055 | myotome development(GO:0061055) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.2 | 0.9 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.2 | 0.5 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.2 | 0.5 | GO:0036071 | N-glycan fucosylation(GO:0036071) |
| 0.2 | 1.8 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.1 | 0.4 | GO:0100009 | regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.1 | 0.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 1.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.5 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 1.7 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 0.3 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.1 | 4.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 1.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.2 | GO:0072019 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.1 | 1.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.8 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.6 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.4 | GO:0035624 | receptor transactivation(GO:0035624) |
| 0.1 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.1 | 3.9 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.3 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.0 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.7 | GO:0044130 | negative regulation of growth of symbiont in host(GO:0044130) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.6 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.4 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.9 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 1.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0061083 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.9 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.8 | GO:0007585 | respiratory gaseous exchange(GO:0007585) |
| 0.0 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.3 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0021779 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 2.9 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.0 | GO:0048617 | foregut morphogenesis(GO:0007440) embryonic foregut morphogenesis(GO:0048617) |
| 0.0 | 0.1 | GO:2000782 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.3 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0045023 | G0 to G1 transition(GO:0045023) |
| 0.0 | 0.3 | GO:0044419 | symbiosis, encompassing mutualism through parasitism(GO:0044403) interspecies interaction between organisms(GO:0044419) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.0 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.0 | GO:0048852 | hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) |
| 0.0 | 0.2 | GO:0003094 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.1 | GO:0034312 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 6.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.3 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 1.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 2.1 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.2 | 1.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.2 | 1.8 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 3.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 3.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 1.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 0.3 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.2 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.6 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0033597 | mitotic checkpoint complex(GO:0033597) bub1-bub3 complex(GO:1990298) |
| 0.0 | 0.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.1 | GO:0035578 | azurophil granule lumen(GO:0035578) |