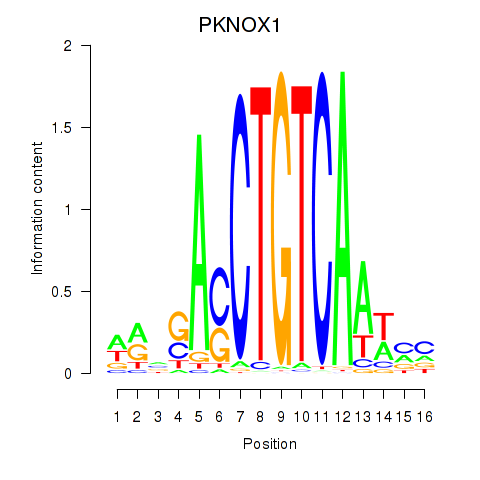
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for PKNOX1_TGIF2
Z-value: 1.50
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PKNOX1 |
|
TGIF2
|
ENSG00000118707.5 | TGIF2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| TGIF2 | hg19_v2_chr20_+_35201993_35202050, hg19_v2_chr20_+_35201857_35201891 | 0.75 | 9.0e-04 | Click! |
| PKNOX1 | hg19_v2_chr21_+_44394620_44394737 | -0.49 | 5.6e-02 | Click! |
Activity profile of PKNOX1_TGIF2 motif
Sorted Z-values of PKNOX1_TGIF2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PKNOX1_TGIF2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_52828805 | 5.56 |
ENST00000416872.2 ENST00000449956.2 |
ITIH3 |
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr1_+_207262540 | 3.96 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr1_+_207262881 | 3.93 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr1_+_207262170 | 3.59 |
ENST00000367078.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr4_+_166300084 | 3.52 |
ENST00000402744.4 |
CPE |
carboxypeptidase E |
| chr1_+_207262578 | 3.52 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr1_+_207262627 | 3.49 |
ENST00000391923.1 |
C4BPB |
complement component 4 binding protein, beta |
| chr2_+_118846008 | 3.42 |
ENST00000245787.4 |
INSIG2 |
insulin induced gene 2 |
| chr6_+_138188551 | 3.07 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr14_-_94856987 | 2.89 |
ENST00000449399.3 ENST00000404814.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr14_-_94857004 | 2.79 |
ENST00000557492.1 ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr14_-_94856951 | 2.79 |
ENST00000553327.1 ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr17_-_64225508 | 2.43 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr21_-_34186006 | 2.28 |
ENST00000490358.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr9_-_33447584 | 2.15 |
ENST00000297991.4 |
AQP3 |
aquaporin 3 (Gill blood group) |
| chr10_-_118764862 | 2.08 |
ENST00000260777.10 |
KIAA1598 |
KIAA1598 |
| chr21_-_34185944 | 2.08 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr15_-_61521495 | 2.03 |
ENST00000335670.6 |
RORA |
RAR-related orphan receptor A |
| chr21_-_34185989 | 1.96 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr6_+_111195973 | 1.95 |
ENST00000368885.3 ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1 |
adenosylmethionine decarboxylase 1 |
| chr7_+_29234028 | 1.81 |
ENST00000222792.6 |
CHN2 |
chimerin 2 |
| chr6_+_26124373 | 1.81 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr16_-_30457048 | 1.79 |
ENST00000500504.2 ENST00000542752.1 |
SEPHS2 |
selenophosphate synthetase 2 |
| chr16_+_2588012 | 1.75 |
ENST00000354836.5 ENST00000389224.3 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr8_-_17752912 | 1.71 |
ENST00000398054.1 ENST00000381840.2 |
FGL1 |
fibrinogen-like 1 |
| chr6_-_32140886 | 1.68 |
ENST00000395496.1 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr7_+_29234101 | 1.61 |
ENST00000435288.2 |
CHN2 |
chimerin 2 |
| chr9_-_111696340 | 1.60 |
ENST00000374647.5 |
IKBKAP |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr16_+_30212378 | 1.58 |
ENST00000569485.1 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr7_-_148580563 | 1.53 |
ENST00000476773.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chr10_-_73848531 | 1.50 |
ENST00000373109.2 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr22_+_17082732 | 1.50 |
ENST00000558085.2 ENST00000592918.1 ENST00000400593.2 ENST00000592107.1 ENST00000426585.1 ENST00000591299.1 |
TPTEP1 |
transmembrane phosphatase with tensin homology pseudogene 1 |
| chr11_-_107729887 | 1.47 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr7_-_156803329 | 1.44 |
ENST00000252971.6 |
MNX1 |
motor neuron and pancreas homeobox 1 |
| chr13_-_41837620 | 1.32 |
ENST00000379477.1 ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1 |
mitochondrial translational release factor 1 |
| chr10_-_11574274 | 1.30 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr1_-_63988846 | 1.29 |
ENST00000283568.8 ENST00000371092.3 ENST00000271002.10 |
ITGB3BP |
integrin beta 3 binding protein (beta3-endonexin) |
| chr1_+_2036149 | 1.27 |
ENST00000482686.1 ENST00000400920.1 ENST00000486681.1 |
PRKCZ |
protein kinase C, zeta |
| chr3_-_142166904 | 1.25 |
ENST00000264951.4 |
XRN1 |
5'-3' exoribonuclease 1 |
| chr1_-_54872059 | 1.22 |
ENST00000371320.3 |
SSBP3 |
single stranded DNA binding protein 3 |
| chr3_-_49726486 | 1.22 |
ENST00000449682.2 |
MST1 |
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr3_+_52719936 | 1.17 |
ENST00000418458.1 ENST00000394799.2 |
GNL3 |
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr10_-_73848764 | 1.15 |
ENST00000317376.4 ENST00000412663.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr4_-_175443943 | 1.15 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr1_+_22778337 | 1.12 |
ENST00000404138.1 ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40 |
zinc finger and BTB domain containing 40 |
| chr1_+_154966058 | 1.09 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr16_-_3030407 | 1.06 |
ENST00000431515.2 ENST00000574385.1 ENST00000576268.1 ENST00000574730.1 ENST00000575632.1 ENST00000573944.1 ENST00000262300.8 |
PKMYT1 |
protein kinase, membrane associated tyrosine/threonine 1 |
| chr19_-_12512062 | 1.05 |
ENST00000595766.1 ENST00000430385.3 |
ZNF799 |
zinc finger protein 799 |
| chr14_-_21493123 | 1.05 |
ENST00000556147.1 ENST00000554489.1 ENST00000555657.1 ENST00000557274.1 ENST00000555158.1 ENST00000554833.1 ENST00000555384.1 ENST00000556420.1 ENST00000554893.1 ENST00000553503.1 ENST00000555733.1 ENST00000553867.1 ENST00000397856.3 ENST00000397855.3 ENST00000556008.1 ENST00000557182.1 ENST00000554483.1 ENST00000556688.1 ENST00000397853.3 ENST00000556329.2 ENST00000554143.1 ENST00000397851.2 ENST00000555142.1 ENST00000557676.1 ENST00000556924.1 |
NDRG2 |
NDRG family member 2 |
| chr2_+_85804614 | 1.04 |
ENST00000263864.5 ENST00000409760.1 |
VAMP8 |
vesicle-associated membrane protein 8 |
| chr5_-_169725231 | 1.03 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr13_+_33160553 | 1.02 |
ENST00000315596.10 |
PDS5B |
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr6_+_31865552 | 1.02 |
ENST00000469372.1 ENST00000497706.1 |
C2 |
complement component 2 |
| chr7_+_94536898 | 1.00 |
ENST00000433360.1 ENST00000340694.4 ENST00000424654.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr14_-_94759408 | 0.99 |
ENST00000554723.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr2_+_242127924 | 0.99 |
ENST00000402530.3 ENST00000274979.8 ENST00000402430.3 |
ANO7 |
anoctamin 7 |
| chr2_+_233925064 | 0.98 |
ENST00000359570.5 ENST00000538935.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr10_-_118765081 | 0.98 |
ENST00000392903.2 ENST00000355371.4 |
KIAA1598 |
KIAA1598 |
| chr18_-_52626622 | 0.98 |
ENST00000591504.1 |
CCDC68 |
coiled-coil domain containing 68 |
| chr14_-_94759361 | 0.96 |
ENST00000393096.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr19_-_39322299 | 0.96 |
ENST00000601094.1 ENST00000595567.1 ENST00000602115.1 ENST00000601778.1 ENST00000597205.1 ENST00000595470.1 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chr2_+_113885138 | 0.94 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr1_-_247335269 | 0.92 |
ENST00000543802.2 ENST00000491356.1 ENST00000472531.1 ENST00000340684.6 |
ZNF124 |
zinc finger protein 124 |
| chr17_+_38296576 | 0.89 |
ENST00000264645.7 |
CASC3 |
cancer susceptibility candidate 3 |
| chr10_+_28966271 | 0.88 |
ENST00000375533.3 |
BAMBI |
BMP and activin membrane-bound inhibitor |
| chr8_+_11666649 | 0.88 |
ENST00000528643.1 ENST00000525777.1 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr12_-_31479045 | 0.86 |
ENST00000539409.1 ENST00000395766.1 |
FAM60A |
family with sequence similarity 60, member A |
| chr1_+_28844648 | 0.86 |
ENST00000373832.1 ENST00000373831.3 |
RCC1 |
regulator of chromosome condensation 1 |
| chr13_-_46716969 | 0.85 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr4_-_105416039 | 0.83 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr19_-_12551849 | 0.82 |
ENST00000595562.1 ENST00000301547.5 |
CTD-3105H18.16 ZNF443 |
Uncharacterized protein zinc finger protein 443 |
| chr3_-_3221358 | 0.79 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr10_+_28822636 | 0.78 |
ENST00000442148.1 ENST00000448193.1 |
WAC |
WW domain containing adaptor with coiled-coil |
| chr8_-_141774467 | 0.75 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr5_-_58882219 | 0.75 |
ENST00000505453.1 ENST00000360047.5 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr15_-_37392086 | 0.74 |
ENST00000561208.1 |
MEIS2 |
Meis homeobox 2 |
| chr4_+_3076388 | 0.72 |
ENST00000355072.5 |
HTT |
huntingtin |
| chr2_+_62900986 | 0.71 |
ENST00000405015.3 ENST00000413434.1 ENST00000426940.1 ENST00000449820.1 |
EHBP1 |
EH domain binding protein 1 |
| chr10_+_114710516 | 0.71 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_2867325 | 0.70 |
ENST00000307635.2 ENST00000586426.1 |
ZNF556 |
zinc finger protein 556 |
| chr16_+_2587998 | 0.70 |
ENST00000441549.3 ENST00000268673.7 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr15_+_68346501 | 0.68 |
ENST00000249636.6 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
| chr1_+_74701062 | 0.68 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr19_-_39322497 | 0.66 |
ENST00000221418.4 |
ECH1 |
enoyl CoA hydratase 1, peroxisomal |
| chr14_-_21492113 | 0.66 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr8_-_99837856 | 0.65 |
ENST00000518165.1 ENST00000419617.2 |
STK3 |
serine/threonine kinase 3 |
| chr19_+_49496705 | 0.65 |
ENST00000595090.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr9_-_132805430 | 0.65 |
ENST00000446176.2 ENST00000355681.3 ENST00000420781.1 |
FNBP1 |
formin binding protein 1 |
| chrX_+_135730373 | 0.64 |
ENST00000370628.2 |
CD40LG |
CD40 ligand |
| chr10_+_51565188 | 0.64 |
ENST00000430396.2 ENST00000374087.4 ENST00000414907.2 |
NCOA4 |
nuclear receptor coactivator 4 |
| chr21_-_34143971 | 0.62 |
ENST00000290178.4 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
| chrX_+_135730297 | 0.62 |
ENST00000370629.2 |
CD40LG |
CD40 ligand |
| chr2_-_24149977 | 0.61 |
ENST00000238789.5 |
ATAD2B |
ATPase family, AAA domain containing 2B |
| chr7_+_29234375 | 0.60 |
ENST00000409350.1 ENST00000495789.2 ENST00000539389.1 |
CHN2 |
chimerin 2 |
| chr12_+_72148614 | 0.60 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr12_-_91451758 | 0.59 |
ENST00000266719.3 |
KERA |
keratocan |
| chr2_+_128848881 | 0.56 |
ENST00000259253.6 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr9_-_124989804 | 0.56 |
ENST00000373755.2 ENST00000373754.2 |
LHX6 |
LIM homeobox 6 |
| chr1_+_205473720 | 0.56 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr14_-_21492251 | 0.55 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr2_-_183106641 | 0.55 |
ENST00000346717.4 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr3_+_111260954 | 0.55 |
ENST00000283285.5 |
CD96 |
CD96 molecule |
| chr3_+_185080908 | 0.54 |
ENST00000265026.3 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chr9_+_133454943 | 0.54 |
ENST00000319725.9 |
FUBP3 |
far upstream element (FUSE) binding protein 3 |
| chr1_-_225615599 | 0.53 |
ENST00000421383.1 ENST00000272163.4 |
LBR |
lamin B receptor |
| chr4_-_46391805 | 0.53 |
ENST00000540012.1 |
GABRA2 |
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr1_-_53163992 | 0.53 |
ENST00000371538.3 |
SELRC1 |
cytochrome c oxidase assembly factor 7 |
| chr3_-_52569023 | 0.52 |
ENST00000307076.4 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
| chr19_-_10764509 | 0.51 |
ENST00000591501.1 |
ILF3-AS1 |
ILF3 antisense RNA 1 (head to head) |
| chr2_+_128848740 | 0.51 |
ENST00000375990.3 |
UGGT1 |
UDP-glucose glycoprotein glucosyltransferase 1 |
| chr4_-_83483395 | 0.49 |
ENST00000515780.2 |
TMEM150C |
transmembrane protein 150C |
| chr19_-_45663408 | 0.49 |
ENST00000317951.4 |
NKPD1 |
NTPase, KAP family P-loop domain containing 1 |
| chr19_-_13044494 | 0.49 |
ENST00000593021.1 ENST00000587981.1 ENST00000423140.2 ENST00000314606.4 |
FARSA |
phenylalanyl-tRNA synthetase, alpha subunit |
| chr3_+_63897605 | 0.48 |
ENST00000487717.1 |
ATXN7 |
ataxin 7 |
| chr1_-_51425902 | 0.48 |
ENST00000396153.2 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr20_-_33872518 | 0.47 |
ENST00000374436.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr7_+_95115210 | 0.47 |
ENST00000428113.1 ENST00000325885.5 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
| chr12_-_91398796 | 0.47 |
ENST00000261172.3 ENST00000551767.1 |
EPYC |
epiphycan |
| chr2_+_99953816 | 0.46 |
ENST00000289371.6 |
EIF5B |
eukaryotic translation initiation factor 5B |
| chr17_+_13972807 | 0.46 |
ENST00000429152.2 ENST00000261643.3 ENST00000536205.1 ENST00000537334.1 |
COX10 |
cytochrome c oxidase assembly homolog 10 (yeast) |
| chr6_-_131949200 | 0.44 |
ENST00000539158.1 ENST00000368058.1 |
MED23 |
mediator complex subunit 23 |
| chr7_-_104909435 | 0.44 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr18_+_3449821 | 0.43 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr6_-_35656685 | 0.43 |
ENST00000539068.1 ENST00000540787.1 |
FKBP5 |
FK506 binding protein 5 |
| chr19_+_49496782 | 0.43 |
ENST00000601968.1 ENST00000596837.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr11_+_129245796 | 0.41 |
ENST00000281437.4 |
BARX2 |
BARX homeobox 2 |
| chr20_-_33872548 | 0.40 |
ENST00000374443.3 |
EIF6 |
eukaryotic translation initiation factor 6 |
| chr21_-_34144157 | 0.40 |
ENST00000331923.4 |
PAXBP1 |
PAX3 and PAX7 binding protein 1 |
| chr21_-_43735446 | 0.38 |
ENST00000398431.2 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr6_-_86099898 | 0.38 |
ENST00000455071.1 |
RP11-30P6.6 |
RP11-30P6.6 |
| chr17_-_39661849 | 0.37 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr19_-_51538148 | 0.37 |
ENST00000319590.4 ENST00000250351.4 |
KLK12 |
kallikrein-related peptidase 12 |
| chr2_-_61765315 | 0.36 |
ENST00000406957.1 ENST00000401558.2 |
XPO1 |
exportin 1 (CRM1 homolog, yeast) |
| chr8_-_17555164 | 0.35 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr5_-_132073111 | 0.34 |
ENST00000403231.1 |
KIF3A |
kinesin family member 3A |
| chr14_-_21994525 | 0.34 |
ENST00000538754.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr19_-_51538118 | 0.34 |
ENST00000529888.1 |
KLK12 |
kallikrein-related peptidase 12 |
| chr12_+_69201923 | 0.34 |
ENST00000462284.1 ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr16_+_16484691 | 0.34 |
ENST00000344087.4 |
NPIPA7 |
nuclear pore complex interacting protein family, member A7 |
| chr17_+_76374714 | 0.33 |
ENST00000262764.6 ENST00000589689.1 ENST00000329897.7 ENST00000592043.1 ENST00000587356.1 |
PGS1 |
phosphatidylglycerophosphate synthase 1 |
| chr19_-_12267524 | 0.32 |
ENST00000455799.1 ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625 |
zinc finger protein 625 |
| chr16_+_2587965 | 0.32 |
ENST00000342085.4 ENST00000566659.1 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr2_-_152589670 | 0.32 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chr6_+_41021027 | 0.31 |
ENST00000244669.2 |
APOBEC2 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 |
| chr6_-_131949305 | 0.30 |
ENST00000368053.4 ENST00000354577.4 ENST00000403834.3 ENST00000540546.1 ENST00000368068.3 ENST00000368060.3 |
MED23 |
mediator complex subunit 23 |
| chr5_+_71014990 | 0.30 |
ENST00000296777.4 |
CARTPT |
CART prepropeptide |
| chr6_-_28220002 | 0.30 |
ENST00000377294.2 |
ZKSCAN4 |
zinc finger with KRAB and SCAN domains 4 |
| chr6_-_163148780 | 0.30 |
ENST00000366892.1 ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2 |
parkin RBR E3 ubiquitin protein ligase |
| chr15_+_51200871 | 0.30 |
ENST00000560508.1 |
AP4E1 |
adaptor-related protein complex 4, epsilon 1 subunit |
| chr22_+_50946645 | 0.29 |
ENST00000420993.2 ENST00000395698.3 ENST00000395701.3 ENST00000523045.1 ENST00000299821.11 |
NCAPH2 |
non-SMC condensin II complex, subunit H2 |
| chr5_+_173763250 | 0.29 |
ENST00000515513.1 ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1 |
RP11-267A15.1 |
| chr3_+_111260856 | 0.29 |
ENST00000352690.4 |
CD96 |
CD96 molecule |
| chr4_+_57774042 | 0.29 |
ENST00000309042.7 |
REST |
RE1-silencing transcription factor |
| chr16_-_30569584 | 0.28 |
ENST00000252797.2 ENST00000568114.1 |
ZNF764 AC002310.13 |
zinc finger protein 764 Uncharacterized protein |
| chr4_-_46391367 | 0.28 |
ENST00000503806.1 ENST00000356504.1 ENST00000514090.1 ENST00000506961.1 |
GABRA2 |
gamma-aminobutyric acid (GABA) A receptor, alpha 2 |
| chr7_+_76139833 | 0.27 |
ENST00000257632.5 |
UPK3B |
uroplakin 3B |
| chr11_-_111741994 | 0.27 |
ENST00000398006.2 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
| chr13_+_24844819 | 0.27 |
ENST00000399949.2 |
SPATA13 |
spermatogenesis associated 13 |
| chr6_+_112375462 | 0.27 |
ENST00000361714.1 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
| chr3_+_38017264 | 0.27 |
ENST00000436654.1 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr1_+_24286287 | 0.27 |
ENST00000334351.7 ENST00000374468.1 |
PNRC2 |
proline-rich nuclear receptor coactivator 2 |
| chr18_+_3451646 | 0.27 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr3_-_192445289 | 0.26 |
ENST00000430714.1 ENST00000418610.1 ENST00000448795.1 ENST00000445105.2 |
FGF12 |
fibroblast growth factor 12 |
| chr16_-_20817753 | 0.26 |
ENST00000389345.5 ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2 |
ERI1 exoribonuclease family member 2 |
| chr19_+_49497121 | 0.26 |
ENST00000413176.2 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr21_-_10990830 | 0.25 |
ENST00000361285.4 ENST00000342420.5 ENST00000328758.5 |
TPTE |
transmembrane phosphatase with tensin homology |
| chr20_+_60878005 | 0.25 |
ENST00000253003.2 |
ADRM1 |
adhesion regulating molecule 1 |
| chr6_+_110299501 | 0.25 |
ENST00000414000.2 |
GPR6 |
G protein-coupled receptor 6 |
| chr12_+_10658489 | 0.25 |
ENST00000538173.1 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr2_+_233271546 | 0.24 |
ENST00000295453.3 |
ALPPL2 |
alkaline phosphatase, placental-like 2 |
| chr22_-_50219548 | 0.24 |
ENST00000404034.1 |
BRD1 |
bromodomain containing 1 |
| chr2_+_16080659 | 0.24 |
ENST00000281043.3 |
MYCN |
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr17_-_29151794 | 0.24 |
ENST00000324238.6 |
CRLF3 |
cytokine receptor-like factor 3 |
| chr8_+_97506033 | 0.23 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr21_-_10990888 | 0.23 |
ENST00000298232.7 |
TPTE |
transmembrane phosphatase with tensin homology |
| chr1_-_32384693 | 0.23 |
ENST00000602683.1 ENST00000470404.1 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
| chr17_-_3499125 | 0.23 |
ENST00000399759.3 |
TRPV1 |
transient receptor potential cation channel, subfamily V, member 1 |
| chr17_-_40337470 | 0.23 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr5_-_154230130 | 0.23 |
ENST00000519501.1 ENST00000518651.1 ENST00000517938.1 ENST00000520461.1 |
FAXDC2 |
fatty acid hydroxylase domain containing 2 |
| chr3_+_49726932 | 0.23 |
ENST00000327697.6 ENST00000432042.1 ENST00000454491.1 |
RNF123 |
ring finger protein 123 |
| chr12_-_30887948 | 0.22 |
ENST00000433722.2 |
CAPRIN2 |
caprin family member 2 |
| chr19_-_6424783 | 0.22 |
ENST00000398148.3 |
KHSRP |
KH-type splicing regulatory protein |
| chr1_+_171217677 | 0.22 |
ENST00000402921.2 |
FMO1 |
flavin containing monooxygenase 1 |
| chr6_-_26027480 | 0.22 |
ENST00000377364.3 |
HIST1H4B |
histone cluster 1, H4b |
| chr12_-_104531785 | 0.22 |
ENST00000551727.1 |
NFYB |
nuclear transcription factor Y, beta |
| chr10_-_124768300 | 0.22 |
ENST00000368886.5 |
IKZF5 |
IKAROS family zinc finger 5 (Pegasus) |
| chr22_-_30234218 | 0.22 |
ENST00000307790.3 ENST00000542393.1 ENST00000397771.2 |
ASCC2 |
activating signal cointegrator 1 complex subunit 2 |
| chr4_+_128886532 | 0.22 |
ENST00000444616.1 ENST00000388795.5 |
C4orf29 |
chromosome 4 open reading frame 29 |
| chr1_+_26348259 | 0.22 |
ENST00000374280.3 |
EXTL1 |
exostosin-like glycosyltransferase 1 |
| chr11_-_84028180 | 0.21 |
ENST00000280241.8 |
DLG2 |
discs, large homolog 2 (Drosophila) |
| chr3_-_183273477 | 0.21 |
ENST00000341319.3 |
KLHL6 |
kelch-like family member 6 |
| chr15_-_43559055 | 0.21 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chr15_-_37391614 | 0.21 |
ENST00000219869.9 |
MEIS2 |
Meis homeobox 2 |
| chr8_-_144241664 | 0.20 |
ENST00000342752.4 |
LY6H |
lymphocyte antigen 6 complex, locus H |
| chrX_+_101380642 | 0.20 |
ENST00000372780.1 ENST00000329035.2 |
TCEAL2 |
transcription elongation factor A (SII)-like 2 |
| chr7_-_99332719 | 0.20 |
ENST00000336374.2 |
CYP3A7 |
cytochrome P450, family 3, subfamily A, polypeptide 7 |
| chr2_-_37068530 | 0.20 |
ENST00000593798.1 |
AC007382.1 |
Uncharacterized protein |
| chr1_-_33502528 | 0.20 |
ENST00000354858.6 |
AK2 |
adenylate kinase 2 |
| chr13_+_73302047 | 0.20 |
ENST00000377814.2 ENST00000377815.3 ENST00000390667.5 |
BORA |
bora, aurora kinase A activator |
| chr5_-_58571935 | 0.20 |
ENST00000503258.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.4 | 7.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.2 | 1.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 1.6 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.2 | 18.8 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.2 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.0 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.1 | 1.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.7 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 1.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.1 | 8.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.3 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.5 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 1.1 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 0.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 1.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 3.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 3.3 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.1 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.3 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 26.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 3.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.8 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 7.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 2.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 2.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.3 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.5 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 1.5 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 19.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 2.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 3.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 1.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 2.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.0 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.1 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 1.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 7.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 2.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.3 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 18.5 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 1.0 | 3.1 | GO:0034146 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.7 | 3.5 | GO:0030070 | insulin processing(GO:0030070) |
| 0.7 | 2.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.6 | 1.8 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.5 | 1.5 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.4 | 2.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.4 | 1.2 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.4 | 1.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.3 | 1.0 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.3 | 1.3 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 1.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.3 | 2.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.3 | 0.8 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.3 | 2.8 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.3 | 2.9 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.3 | 1.0 | GO:1904048 | regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.2 | 1.6 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.2 | 2.4 | GO:0051918 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) negative regulation of fibrinolysis(GO:0051918) |
| 0.2 | 0.9 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 | 1.3 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.8 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.2 | 0.7 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.2 | 1.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 1.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.2 | 0.9 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.2 | 1.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.2 | 2.3 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 1.3 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.1 | 1.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 2.0 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.7 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 2.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 1.9 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.4 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 0.7 | GO:0044334 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.1 | 0.9 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.3 | GO:0071301 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 1.7 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.1 | 7.6 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.9 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 5.6 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 1.3 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.3 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 1.0 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.1 | 1.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 1.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.3 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 1.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.9 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 1.3 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.1 | 0.8 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 0.6 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 | 0.3 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.6 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 1.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.9 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.1 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.9 | GO:0030336 | negative regulation of cell migration(GO:0030336) negative regulation of cell motility(GO:2000146) |
| 0.0 | 0.6 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.4 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.5 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 1.3 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.3 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.5 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.3 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.0 | 0.2 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.9 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.2 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.6 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 1.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.2 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.0 | 0.2 | GO:0010629 | negative regulation of gene expression(GO:0010629) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 0.3 | GO:0021915 | neural tube development(GO:0021915) |
| 0.0 | 1.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:0070268 | cornification(GO:0070268) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.6 | 1.8 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.5 | 2.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.5 | 2.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.4 | 1.3 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.4 | 1.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.3 | 1.3 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.3 | 1.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.3 | 0.9 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.3 | 1.9 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 1.6 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.3 | 3.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.2 | 1.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 1.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.2 | 0.9 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.2 | 0.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 0.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.2 | 1.6 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.2 | 1.2 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.1 | 16.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.9 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 2.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.3 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 1.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 1.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.9 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.1 | 0.5 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 1.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 1.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.8 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 0.2 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.8 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.4 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.7 | GO:0070016 | gamma-catenin binding(GO:0045295) armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.4 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 2.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 3.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 1.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.8 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.3 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.6 | GO:0016853 | isomerase activity(GO:0016853) |