Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
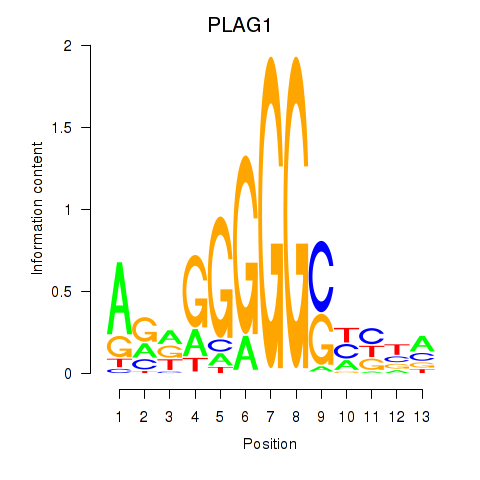
Results for PLAG1
Z-value: 1.14
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.3 | PLAG1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PLAG1 | hg19_v2_chr8_-_57123815_57123867 | 0.55 | 2.8e-02 | Click! |
Activity profile of PLAG1 motif
Sorted Z-values of PLAG1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PLAG1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_51927919 | 4.41 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chr11_-_2160180 | 4.35 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr14_-_25519317 | 3.17 |
ENST00000323944.5 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr14_-_25519095 | 2.87 |
ENST00000419632.2 ENST00000358326.2 ENST00000396700.1 ENST00000548724.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr2_-_21266935 | 2.64 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chrY_+_22737604 | 2.10 |
ENST00000361365.2 |
EIF1AY |
eukaryotic translation initiation factor 1A, Y-linked |
| chr1_-_31845914 | 1.94 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr9_+_139874683 | 1.86 |
ENST00000444903.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr20_+_35169885 | 1.75 |
ENST00000279022.2 ENST00000346786.2 |
MYL9 |
myosin, light chain 9, regulatory |
| chr12_+_57522258 | 1.68 |
ENST00000553277.1 ENST00000243077.3 |
LRP1 |
low density lipoprotein receptor-related protein 1 |
| chr19_+_41725088 | 1.66 |
ENST00000301178.4 |
AXL |
AXL receptor tyrosine kinase |
| chr4_-_155533787 | 1.60 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr16_+_31483374 | 1.56 |
ENST00000394863.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr16_+_31483451 | 1.49 |
ENST00000565360.1 ENST00000361773.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr12_-_106641728 | 1.47 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr1_-_23886285 | 1.15 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr16_-_65155833 | 1.02 |
ENST00000566827.1 ENST00000394156.3 ENST00000562998.1 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr16_-_29910853 | 1.01 |
ENST00000308713.5 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr22_-_30642782 | 0.93 |
ENST00000249075.3 |
LIF |
leukemia inhibitory factor |
| chr1_+_163038565 | 0.90 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr11_+_46299199 | 0.88 |
ENST00000529193.1 ENST00000288400.3 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
| chr4_+_55095264 | 0.87 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_-_85788652 | 0.85 |
ENST00000430215.3 |
GGCX |
gamma-glutamyl carboxylase |
| chr14_-_75079026 | 0.84 |
ENST00000261978.4 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
| chr2_-_86850949 | 0.80 |
ENST00000237455.4 |
RNF103 |
ring finger protein 103 |
| chr12_+_56546363 | 0.79 |
ENST00000551834.1 ENST00000552568.1 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr6_-_43021437 | 0.79 |
ENST00000265348.3 |
CUL7 |
cullin 7 |
| chr16_-_30134524 | 0.79 |
ENST00000395202.1 ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr14_+_32546485 | 0.79 |
ENST00000345122.3 ENST00000432921.1 ENST00000433497.1 |
ARHGAP5 |
Rho GTPase activating protein 5 |
| chr17_-_79805146 | 0.77 |
ENST00000415593.1 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr3_-_73673991 | 0.75 |
ENST00000308537.4 ENST00000263666.4 |
PDZRN3 |
PDZ domain containing ring finger 3 |
| chr1_-_20306909 | 0.75 |
ENST00000375111.3 ENST00000400520.3 |
PLA2G2A |
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr16_-_29910365 | 0.74 |
ENST00000346932.5 ENST00000350527.3 ENST00000537485.1 ENST00000568380.1 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
| chr6_-_43021612 | 0.70 |
ENST00000535468.1 |
CUL7 |
cullin 7 |
| chr14_+_79745746 | 0.70 |
ENST00000281127.7 |
NRXN3 |
neurexin 3 |
| chr17_-_56065484 | 0.68 |
ENST00000581208.1 |
VEZF1 |
vascular endothelial zinc finger 1 |
| chr7_+_72742162 | 0.68 |
ENST00000431982.2 |
FKBP6 |
FK506 binding protein 6, 36kDa |
| chr6_+_43044003 | 0.67 |
ENST00000230419.4 ENST00000476760.1 ENST00000471863.1 ENST00000349241.2 ENST00000352931.2 ENST00000345201.2 |
PTK7 |
protein tyrosine kinase 7 |
| chr16_-_18573396 | 0.65 |
ENST00000543392.1 ENST00000381474.3 ENST00000330537.6 |
NOMO2 |
NODAL modulator 2 |
| chr2_-_128399706 | 0.65 |
ENST00000426981.1 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
| chr3_+_124303539 | 0.65 |
ENST00000428018.2 |
KALRN |
kalirin, RhoGEF kinase |
| chr10_+_111767720 | 0.64 |
ENST00000356080.4 ENST00000277900.8 |
ADD3 |
adducin 3 (gamma) |
| chr17_-_3599327 | 0.59 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr7_-_158380371 | 0.58 |
ENST00000389418.4 ENST00000389416.4 |
PTPRN2 |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr7_-_158380465 | 0.56 |
ENST00000389413.3 ENST00000409483.1 |
PTPRN2 |
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr11_-_73309228 | 0.54 |
ENST00000356467.4 ENST00000064778.4 |
FAM168A |
family with sequence similarity 168, member A |
| chr3_-_52001448 | 0.54 |
ENST00000461554.1 ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chr9_+_34652164 | 0.53 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr7_-_22233442 | 0.53 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr8_-_23261589 | 0.51 |
ENST00000524168.1 ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2 |
lysyl oxidase-like 2 |
| chr7_+_72742178 | 0.50 |
ENST00000442793.1 ENST00000413573.2 ENST00000252037.4 |
FKBP6 |
FK506 binding protein 6, 36kDa |
| chr16_+_29984962 | 0.50 |
ENST00000308893.4 |
TAOK2 |
TAO kinase 2 |
| chr5_+_75699149 | 0.49 |
ENST00000379730.3 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chr14_+_79745682 | 0.48 |
ENST00000557594.1 |
NRXN3 |
neurexin 3 |
| chr21_-_15755446 | 0.47 |
ENST00000544452.1 ENST00000285667.3 |
HSPA13 |
heat shock protein 70kDa family, member 13 |
| chr19_+_41725140 | 0.46 |
ENST00000359092.3 |
AXL |
AXL receptor tyrosine kinase |
| chr20_+_18269121 | 0.45 |
ENST00000377671.3 ENST00000360010.5 ENST00000396026.3 ENST00000402618.2 ENST00000401790.1 ENST00000434018.1 ENST00000538547.1 ENST00000535822.1 |
ZNF133 |
zinc finger protein 133 |
| chr5_-_59189545 | 0.43 |
ENST00000340635.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr10_-_99447024 | 0.43 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr11_+_1940925 | 0.42 |
ENST00000453458.1 ENST00000381557.2 ENST00000381589.3 ENST00000381579.3 ENST00000381563.4 ENST00000344578.4 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
| chr18_-_51750948 | 0.42 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr13_+_53602894 | 0.40 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr8_+_21912328 | 0.37 |
ENST00000432128.1 ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN |
dematin actin binding protein |
| chr11_-_6440283 | 0.37 |
ENST00000299402.6 ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr9_-_135819987 | 0.36 |
ENST00000298552.3 ENST00000403810.1 |
TSC1 |
tuberous sclerosis 1 |
| chr1_-_24469602 | 0.35 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr3_-_58419537 | 0.35 |
ENST00000474765.1 ENST00000485460.1 ENST00000302746.6 ENST00000383714.4 |
PDHB |
pyruvate dehydrogenase (lipoamide) beta |
| chr2_-_74618964 | 0.34 |
ENST00000417090.1 ENST00000409868.1 |
DCTN1 |
dynactin 1 |
| chr2_-_74619152 | 0.34 |
ENST00000440727.1 ENST00000409240.1 |
DCTN1 |
dynactin 1 |
| chr11_-_57282349 | 0.32 |
ENST00000528450.1 |
SLC43A1 |
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr16_-_18812746 | 0.31 |
ENST00000546206.2 ENST00000562819.1 ENST00000562234.2 ENST00000304414.7 ENST00000567078.2 |
ARL6IP1 RP11-1035H13.3 |
ADP-ribosylation factor-like 6 interacting protein 1 Uncharacterized protein |
| chr13_-_49107303 | 0.30 |
ENST00000344532.3 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr17_-_3819751 | 0.28 |
ENST00000225538.3 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
| chr12_-_112819896 | 0.28 |
ENST00000377560.5 ENST00000430131.2 ENST00000550722.1 ENST00000550724.1 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
| chr6_-_150039249 | 0.27 |
ENST00000543571.1 |
LATS1 |
large tumor suppressor kinase 1 |
| chr6_-_31940065 | 0.24 |
ENST00000375349.3 ENST00000337523.5 |
DXO |
decapping exoribonuclease |
| chr5_-_137368708 | 0.24 |
ENST00000033079.3 |
FAM13B |
family with sequence similarity 13, member B |
| chr19_+_45973120 | 0.24 |
ENST00000592811.1 ENST00000586615.1 |
FOSB |
FBJ murine osteosarcoma viral oncogene homolog B |
| chr14_+_79746249 | 0.22 |
ENST00000428277.2 |
NRXN3 |
neurexin 3 |
| chr3_+_32726774 | 0.21 |
ENST00000538368.1 |
CNOT10 |
CCR4-NOT transcription complex, subunit 10 |
| chr10_-_126849588 | 0.21 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chr11_+_63997750 | 0.20 |
ENST00000321685.3 |
DNAJC4 |
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr6_+_31939608 | 0.19 |
ENST00000375331.2 ENST00000375333.2 |
STK19 |
serine/threonine kinase 19 |
| chr7_+_73703728 | 0.18 |
ENST00000361545.5 ENST00000223398.6 |
CLIP2 |
CAP-GLY domain containing linker protein 2 |
| chr22_+_29469100 | 0.17 |
ENST00000327813.5 ENST00000407188.1 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr12_+_107168342 | 0.17 |
ENST00000392837.4 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
| chr1_-_68698222 | 0.17 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr6_-_33285505 | 0.16 |
ENST00000431845.2 |
ZBTB22 |
zinc finger and BTB domain containing 22 |
| chr17_-_4607335 | 0.16 |
ENST00000570571.1 ENST00000575101.1 ENST00000436683.2 ENST00000574876.1 |
PELP1 |
proline, glutamate and leucine rich protein 1 |
| chr1_+_202431859 | 0.13 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr2_-_178937478 | 0.12 |
ENST00000286063.6 |
PDE11A |
phosphodiesterase 11A |
| chr16_+_56485402 | 0.12 |
ENST00000566157.1 ENST00000562150.1 ENST00000561646.1 ENST00000568397.1 |
OGFOD1 |
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr17_-_1588101 | 0.12 |
ENST00000577001.1 ENST00000572621.1 ENST00000304992.6 |
PRPF8 |
pre-mRNA processing factor 8 |
| chr12_-_12419703 | 0.11 |
ENST00000543091.1 ENST00000261349.4 |
LRP6 |
low density lipoprotein receptor-related protein 6 |
| chr9_-_34662651 | 0.11 |
ENST00000259631.4 |
CCL27 |
chemokine (C-C motif) ligand 27 |
| chr4_-_5890145 | 0.10 |
ENST00000397890.2 |
CRMP1 |
collapsin response mediator protein 1 |
| chr20_+_35201857 | 0.10 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr22_+_19744226 | 0.10 |
ENST00000332710.4 ENST00000329705.7 ENST00000359500.3 |
TBX1 |
T-box 1 |
| chr2_-_152955213 | 0.10 |
ENST00000427385.1 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr19_-_2702681 | 0.10 |
ENST00000382159.3 |
GNG7 |
guanine nucleotide binding protein (G protein), gamma 7 |
| chr16_+_31128978 | 0.10 |
ENST00000448516.2 ENST00000219797.4 |
KAT8 |
K(lysine) acetyltransferase 8 |
| chr17_+_38278826 | 0.09 |
ENST00000577454.1 ENST00000578648.1 ENST00000579565.1 |
MSL1 |
male-specific lethal 1 homolog (Drosophila) |
| chr11_-_1643368 | 0.09 |
ENST00000399682.1 |
KRTAP5-4 |
keratin associated protein 5-4 |
| chr16_+_30671223 | 0.08 |
ENST00000568722.1 |
FBRS |
fibrosin |
| chr1_+_22962948 | 0.07 |
ENST00000374642.3 |
C1QA |
complement component 1, q subcomponent, A chain |
| chr10_-_75571566 | 0.07 |
ENST00000299641.4 |
NDST2 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr22_+_29469012 | 0.07 |
ENST00000400335.4 ENST00000400338.2 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr11_-_64511789 | 0.06 |
ENST00000419843.1 ENST00000394430.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr9_-_123476719 | 0.05 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr11_-_2323290 | 0.05 |
ENST00000381153.3 |
C11orf21 |
chromosome 11 open reading frame 21 |
| chr9_+_2621798 | 0.04 |
ENST00000382100.3 |
VLDLR |
very low density lipoprotein receptor |
| chr6_+_31683117 | 0.04 |
ENST00000375825.3 ENST00000375824.1 |
LY6G6D |
lymphocyte antigen 6 complex, locus G6D |
| chr1_-_110613276 | 0.04 |
ENST00000369792.4 |
ALX3 |
ALX homeobox 3 |
| chr9_-_123476612 | 0.04 |
ENST00000426959.1 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr11_-_111170526 | 0.04 |
ENST00000355430.4 |
COLCA1 |
colorectal cancer associated 1 |
| chr12_-_122018859 | 0.03 |
ENST00000536437.1 ENST00000377071.4 ENST00000538046.2 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr8_+_29952914 | 0.03 |
ENST00000321250.8 ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr19_+_38924316 | 0.03 |
ENST00000355481.4 ENST00000360985.3 ENST00000359596.3 |
RYR1 |
ryanodine receptor 1 (skeletal) |
| chr10_-_75571341 | 0.03 |
ENST00000309979.6 |
NDST2 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2 |
| chr4_-_47839966 | 0.02 |
ENST00000273857.4 ENST00000505909.1 ENST00000502252.1 |
CORIN |
corin, serine peptidase |
| chrX_+_23682379 | 0.02 |
ENST00000379349.1 |
PRDX4 |
peroxiredoxin 4 |
| chr1_+_173446405 | 0.02 |
ENST00000340385.5 |
PRDX6 |
peroxiredoxin 6 |
| chr12_-_108733078 | 0.01 |
ENST00000552995.1 ENST00000312143.7 ENST00000397688.2 ENST00000550402.1 |
CMKLR1 |
chemokine-like receptor 1 |
| chr17_-_1420006 | 0.01 |
ENST00000320345.6 ENST00000406424.4 |
INPP5K |
inositol polyphosphate-5-phosphatase K |
| chr1_-_115632035 | 0.01 |
ENST00000433172.1 ENST00000369514.2 ENST00000369516.2 ENST00000369515.2 |
TSPAN2 |
tetraspanin 2 |
| chr5_+_161275320 | 0.00 |
ENST00000437025.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr16_-_46723066 | 0.00 |
ENST00000299138.7 |
VPS35 |
vacuolar protein sorting 35 homolog (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.6 | 1.9 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.5 | 1.9 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.4 | 1.7 | GO:0042954 | apolipoprotein receptor activity(GO:0030226) lipoprotein transporter activity(GO:0042954) |
| 0.3 | 3.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.3 | 6.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.3 | 0.9 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.2 | 3.9 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.8 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.7 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 4.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.4 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 1.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.9 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 0.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 1.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.0 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.4 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 1.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.0 | GO:0038025 | glycoprotein transporter activity(GO:0034437) reelin receptor activity(GO:0038025) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.0 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.6 | 1.9 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.6 | 1.7 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.5 | 4.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.4 | 3.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.4 | 2.6 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.4 | 2.1 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.3 | 0.9 | GO:0043974 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.3 | 0.9 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.2 | 0.8 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.1 | 1.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.6 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.1 | 0.4 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 1.9 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.0 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.1 | 0.9 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 0.8 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.7 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.1 | 1.6 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 1.4 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.1 | 0.7 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.4 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.1 | 1.1 | GO:0030903 | notochord development(GO:0030903) |
| 0.1 | 0.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.4 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.4 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.1 | 0.1 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 0.3 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.1 | 0.3 | GO:0001826 | inner cell mass cell differentiation(GO:0001826) |
| 0.1 | 0.8 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.7 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.3 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.1 | GO:1902809 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.0 | 1.6 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 1.8 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.0 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.7 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.0 | 0.0 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.6 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 0.7 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 1.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 0.0 | GO:0021592 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.0 | 0.7 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) signal transduction involved in mitotic cell cycle checkpoint(GO:0072413) signal transduction involved in mitotic G1 DNA damage checkpoint(GO:0072431) intracellular signal transduction involved in G1 DNA damage checkpoint(GO:1902400) signal transduction involved in mitotic DNA damage checkpoint(GO:1902402) signal transduction involved in mitotic DNA integrity checkpoint(GO:1902403) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 3.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 1.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 0.8 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 2.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 2.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.4 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.2 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 1.5 | GO:1990393 | 3M complex(GO:1990393) |
| 0.3 | 6.0 | GO:0000145 | exocyst(GO:0000145) |
| 0.3 | 0.8 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 2.1 | GO:0033643 | host cell part(GO:0033643) |
| 0.1 | 1.6 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 2.5 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.1 | 1.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.1 | 0.4 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.1 | 0.3 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.9 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 4.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.8 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 2.0 | GO:0016528 | sarcoplasm(GO:0016528) |
| 0.0 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.0 | 1.9 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.0 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.7 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 1.1 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.0 | 0.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 3.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 3.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 2.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 2.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 3.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 2.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |