Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
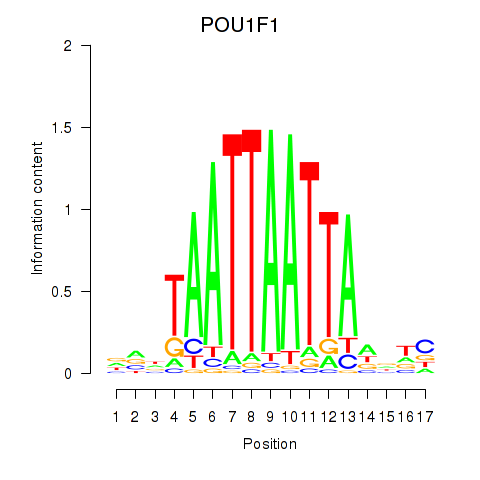
Results for POU1F1
Z-value: 0.69
Transcription factors associated with POU1F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU1F1
|
ENSG00000064835.6 | POU1F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU1F1 | hg19_v2_chr3_-_87325728_87325739 | 0.08 | 7.7e-01 | Click! |
Activity profile of POU1F1 motif
Sorted Z-values of POU1F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU1F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21269404 | 1.88 |
ENST00000313654.9 |
LAMA3 |
laminin, alpha 3 |
| chrX_-_13835147 | 1.72 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr7_+_129932974 | 1.67 |
ENST00000445470.2 ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4 |
carboxypeptidase A4 |
| chr18_+_21269556 | 1.61 |
ENST00000399516.3 |
LAMA3 |
laminin, alpha 3 |
| chr8_-_49833978 | 1.58 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr7_+_134528635 | 1.42 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr8_-_49834299 | 1.36 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr4_+_169552748 | 1.35 |
ENST00000504519.1 ENST00000512127.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr8_+_30244580 | 1.29 |
ENST00000523115.1 ENST00000519647.1 |
RBPMS |
RNA binding protein with multiple splicing |
| chr11_+_12399071 | 1.13 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr5_+_119867159 | 1.08 |
ENST00000505123.1 |
PRR16 |
proline rich 16 |
| chr11_+_35201826 | 1.04 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr10_+_17270214 | 1.02 |
ENST00000544301.1 |
VIM |
vimentin |
| chr2_-_190044480 | 1.02 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr5_+_140762268 | 0.91 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr11_+_2405833 | 0.88 |
ENST00000527343.1 ENST00000464784.2 |
CD81 |
CD81 molecule |
| chr7_+_130126165 | 0.87 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr7_+_130126012 | 0.85 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr7_-_45960850 | 0.84 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chrX_-_106243451 | 0.81 |
ENST00000355610.4 ENST00000535534.1 |
MORC4 |
MORC family CW-type zinc finger 4 |
| chr18_+_29027696 | 0.76 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr10_+_5135981 | 0.75 |
ENST00000380554.3 |
AKR1C3 |
aldo-keto reductase family 1, member C3 |
| chr12_+_26348429 | 0.73 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr21_-_27423339 | 0.72 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr19_-_43969796 | 0.69 |
ENST00000244333.3 |
LYPD3 |
LY6/PLAUR domain containing 3 |
| chr7_+_107224364 | 0.65 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr3_+_138340067 | 0.64 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr17_+_1674982 | 0.64 |
ENST00000572048.1 ENST00000573763.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr12_+_26348246 | 0.62 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr1_+_81771806 | 0.61 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr5_-_125930929 | 0.61 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr11_-_117186946 | 0.61 |
ENST00000313005.6 ENST00000528053.1 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
| chr8_+_98900132 | 0.59 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr1_+_77333117 | 0.58 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chr6_-_110501200 | 0.58 |
ENST00000392586.1 ENST00000419252.1 ENST00000392589.1 ENST00000392588.1 ENST00000359451.2 |
WASF1 |
WAS protein family, member 1 |
| chr11_+_5710919 | 0.56 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr8_+_70404996 | 0.55 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr9_+_125133315 | 0.53 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_+_186990298 | 0.52 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chr17_+_61086917 | 0.51 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr3_-_48130707 | 0.49 |
ENST00000360240.6 ENST00000383737.4 |
MAP4 |
microtubule-associated protein 4 |
| chr6_-_52859046 | 0.48 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr15_-_56757329 | 0.48 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr3_+_138340049 | 0.47 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr10_+_64133934 | 0.45 |
ENST00000395254.3 ENST00000395255.3 ENST00000410046.3 |
ZNF365 |
zinc finger protein 365 |
| chrX_-_102941596 | 0.43 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr10_+_53806501 | 0.41 |
ENST00000373975.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr10_-_75415825 | 0.36 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr6_+_26087646 | 0.36 |
ENST00000309234.6 |
HFE |
hemochromatosis |
| chrX_-_102942961 | 0.35 |
ENST00000434230.1 ENST00000418819.1 ENST00000360458.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chrX_+_43515467 | 0.35 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr1_-_109584608 | 0.34 |
ENST00000400794.3 ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47 |
WD repeat domain 47 |
| chr4_+_3344141 | 0.33 |
ENST00000306648.7 |
RGS12 |
regulator of G-protein signaling 12 |
| chrX_-_102943022 | 0.32 |
ENST00000433176.2 |
MORF4L2 |
mortality factor 4 like 2 |
| chr2_+_161993465 | 0.31 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr10_+_91152303 | 0.31 |
ENST00000371804.3 |
IFIT1 |
interferon-induced protein with tetratricopeptide repeats 1 |
| chr8_-_30670384 | 0.31 |
ENST00000221138.4 ENST00000518243.1 |
PPP2CB |
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr2_+_161993412 | 0.31 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr16_-_66584059 | 0.30 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr10_-_49860525 | 0.30 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr9_-_21351377 | 0.28 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chr14_-_80697396 | 0.28 |
ENST00000557010.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr6_+_26087509 | 0.28 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr12_-_71148413 | 0.28 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr9_+_125132803 | 0.28 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr4_-_122854612 | 0.26 |
ENST00000264811.5 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr12_-_71148357 | 0.26 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr15_+_67418047 | 0.26 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr14_-_25078864 | 0.26 |
ENST00000216338.4 ENST00000557220.2 ENST00000382548.4 |
GZMH |
granzyme H (cathepsin G-like 2, protein h-CCPX) |
| chr15_-_72563585 | 0.25 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr5_+_140593509 | 0.25 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chr16_-_18462221 | 0.23 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr6_+_158733692 | 0.23 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr10_-_75226166 | 0.22 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr11_-_104827425 | 0.21 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr1_+_87012753 | 0.21 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr16_-_66764119 | 0.21 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr9_-_95056010 | 0.21 |
ENST00000443024.2 |
IARS |
isoleucyl-tRNA synthetase |
| chr5_+_115177178 | 0.20 |
ENST00000316788.7 |
AP3S1 |
adaptor-related protein complex 3, sigma 1 subunit |
| chr3_-_27498235 | 0.20 |
ENST00000295736.5 ENST00000428386.1 ENST00000428179.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr1_-_45988542 | 0.20 |
ENST00000424390.1 |
PRDX1 |
peroxiredoxin 1 |
| chr4_+_88754113 | 0.19 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr1_-_197036364 | 0.19 |
ENST00000367412.1 |
F13B |
coagulation factor XIII, B polypeptide |
| chr9_-_33473882 | 0.19 |
ENST00000455041.2 ENST00000353159.2 ENST00000297990.4 ENST00000379471.2 |
NOL6 |
nucleolar protein 6 (RNA-associated) |
| chr8_-_110986918 | 0.19 |
ENST00000297404.1 |
KCNV1 |
potassium channel, subfamily V, member 1 |
| chr22_+_20105259 | 0.19 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr16_+_15489603 | 0.18 |
ENST00000568766.1 ENST00000287594.7 |
RP11-1021N1.1 MPV17L |
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr2_+_210517895 | 0.17 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr20_-_7921090 | 0.17 |
ENST00000378789.3 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr8_-_59412717 | 0.17 |
ENST00000301645.3 |
CYP7A1 |
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr12_-_7656357 | 0.17 |
ENST00000396620.3 ENST00000432237.2 ENST00000359156.4 |
CD163 |
CD163 molecule |
| chr8_+_67687413 | 0.17 |
ENST00000521960.1 ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
| chr16_-_28937027 | 0.16 |
ENST00000358201.4 |
RABEP2 |
rabaptin, RAB GTPase binding effector protein 2 |
| chr3_-_160823158 | 0.16 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr6_-_136788001 | 0.16 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr16_-_70835034 | 0.15 |
ENST00000261776.5 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
| chr1_-_67266939 | 0.15 |
ENST00000304526.2 |
INSL5 |
insulin-like 5 |
| chr11_-_18548426 | 0.15 |
ENST00000357193.3 ENST00000536719.1 |
TSG101 |
tumor susceptibility 101 |
| chr10_-_28571015 | 0.15 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr17_-_40950698 | 0.14 |
ENST00000328434.7 |
COA3 |
cytochrome c oxidase assembly factor 3 |
| chr18_-_64271363 | 0.13 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr12_+_26164645 | 0.13 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr7_-_16872932 | 0.13 |
ENST00000419572.2 ENST00000412973.1 |
AGR2 |
anterior gradient 2 |
| chr1_-_53608249 | 0.13 |
ENST00000371494.4 |
SLC1A7 |
solute carrier family 1 (glutamate transporter), member 7 |
| chr4_-_70518941 | 0.13 |
ENST00000286604.4 ENST00000505512.1 ENST00000514019.1 |
UGT2A1 UGT2A1 |
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr5_+_140552218 | 0.13 |
ENST00000231137.3 |
PCDHB7 |
protocadherin beta 7 |
| chr4_+_74347400 | 0.13 |
ENST00000226355.3 |
AFM |
afamin |
| chr11_+_110300607 | 0.13 |
ENST00000260270.2 |
FDX1 |
ferredoxin 1 |
| chr4_+_190861993 | 0.12 |
ENST00000524583.1 ENST00000531991.2 |
FRG1 |
FSHD region gene 1 |
| chr15_+_84115868 | 0.12 |
ENST00000427482.2 |
SH3GL3 |
SH3-domain GRB2-like 3 |
| chr6_-_49712123 | 0.12 |
ENST00000263045.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr9_-_95055956 | 0.12 |
ENST00000375629.3 ENST00000447699.2 ENST00000375643.3 ENST00000395554.3 |
IARS |
isoleucyl-tRNA synthetase |
| chr4_+_41937131 | 0.12 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr3_-_160823040 | 0.12 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr15_+_64680003 | 0.12 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr3_-_46759314 | 0.11 |
ENST00000315170.7 |
PRSS50 |
protease, serine, 50 |
| chrX_+_154113317 | 0.11 |
ENST00000354461.2 |
H2AFB1 |
H2A histone family, member B1 |
| chr19_+_19144384 | 0.11 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr9_-_100684845 | 0.11 |
ENST00000375119.3 |
C9orf156 |
chromosome 9 open reading frame 156 |
| chr1_-_21377383 | 0.11 |
ENST00000374935.3 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr12_-_10978957 | 0.11 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr16_-_66583701 | 0.10 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr16_+_81069433 | 0.10 |
ENST00000299575.4 |
ATMIN |
ATM interactor |
| chr2_-_26700900 | 0.10 |
ENST00000338581.6 ENST00000339598.3 ENST00000402415.3 |
OTOF |
otoferlin |
| chr19_-_51920952 | 0.10 |
ENST00000356298.5 ENST00000339313.5 ENST00000529627.1 ENST00000439889.2 ENST00000353836.5 ENST00000432469.2 |
SIGLEC10 |
sialic acid binding Ig-like lectin 10 |
| chr7_-_76829125 | 0.10 |
ENST00000248598.5 |
FGL2 |
fibrinogen-like 2 |
| chr11_-_128894053 | 0.10 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr12_-_86650077 | 0.10 |
ENST00000552808.2 ENST00000547225.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr18_-_13915530 | 0.10 |
ENST00000327606.3 |
MC2R |
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr12_-_86650045 | 0.10 |
ENST00000604798.1 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr18_+_616672 | 0.10 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr5_+_140588269 | 0.09 |
ENST00000541609.1 ENST00000239450.2 |
PCDHB12 |
protocadherin beta 12 |
| chr14_-_101295407 | 0.09 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chr5_-_55412774 | 0.09 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr6_-_146057144 | 0.09 |
ENST00000367519.3 |
EPM2A |
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr14_-_78083112 | 0.08 |
ENST00000216484.2 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
| chr6_+_29429217 | 0.08 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr1_-_7913089 | 0.08 |
ENST00000361696.5 |
UTS2 |
urotensin 2 |
| chr6_-_52859968 | 0.08 |
ENST00000370959.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr2_-_231989808 | 0.08 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr22_+_23487513 | 0.08 |
ENST00000263116.2 ENST00000341989.4 |
RAB36 |
RAB36, member RAS oncogene family |
| chr7_-_14026123 | 0.08 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr12_-_67197760 | 0.07 |
ENST00000539540.1 ENST00000540433.1 ENST00000541947.1 ENST00000538373.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chr11_-_102651343 | 0.07 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr4_-_103746683 | 0.07 |
ENST00000504211.1 ENST00000508476.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_145782358 | 0.06 |
ENST00000422482.1 |
AC107021.1 |
HCG1786590; PRO2533; Uncharacterized protein |
| chr1_+_149754227 | 0.06 |
ENST00000444948.1 ENST00000369168.4 |
FCGR1A |
Fc fragment of IgG, high affinity Ia, receptor (CD64) |
| chr12_-_118796910 | 0.06 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr22_-_43010928 | 0.06 |
ENST00000348657.2 ENST00000252115.5 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
| chr15_+_64386261 | 0.06 |
ENST00000560829.1 |
SNX1 |
sorting nexin 1 |
| chr5_+_111755280 | 0.06 |
ENST00000600409.1 |
EPB41L4A-AS2 |
EPB41L4A antisense RNA 2 (head to head) |
| chr15_+_65843130 | 0.06 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr4_+_5526883 | 0.06 |
ENST00000195455.2 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr20_+_3776371 | 0.06 |
ENST00000245960.5 |
CDC25B |
cell division cycle 25B |
| chr3_+_193310918 | 0.06 |
ENST00000361908.3 ENST00000392438.3 ENST00000361510.2 ENST00000361715.2 ENST00000361828.2 ENST00000361150.2 |
OPA1 |
optic atrophy 1 (autosomal dominant) |
| chr1_+_158323755 | 0.06 |
ENST00000368157.1 ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E |
CD1e molecule |
| chr1_+_197237352 | 0.05 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr3_+_136649311 | 0.05 |
ENST00000469404.1 ENST00000467911.1 |
NCK1 |
NCK adaptor protein 1 |
| chr4_-_46996424 | 0.05 |
ENST00000264318.3 |
GABRA4 |
gamma-aminobutyric acid (GABA) A receptor, alpha 4 |
| chrX_-_11308598 | 0.05 |
ENST00000380717.3 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr12_-_10959892 | 0.04 |
ENST00000240615.2 |
TAS2R8 |
taste receptor, type 2, member 8 |
| chr6_-_49712147 | 0.04 |
ENST00000433368.2 ENST00000354620.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr4_-_120243545 | 0.04 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr19_+_21579958 | 0.04 |
ENST00000339914.6 ENST00000599461.1 |
ZNF493 |
zinc finger protein 493 |
| chr3_+_130650738 | 0.04 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_-_54411255 | 0.04 |
ENST00000371377.3 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr8_-_141774467 | 0.04 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr4_+_37455536 | 0.04 |
ENST00000381980.4 ENST00000508175.1 |
C4orf19 |
chromosome 4 open reading frame 19 |
| chr11_+_122709200 | 0.04 |
ENST00000227348.4 |
CRTAM |
cytotoxic and regulatory T cell molecule |
| chr1_+_115572415 | 0.04 |
ENST00000256592.1 |
TSHB |
thyroid stimulating hormone, beta |
| chr3_+_38537960 | 0.03 |
ENST00000453767.1 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
| chr2_-_152118352 | 0.03 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chr9_-_21239978 | 0.03 |
ENST00000380222.2 |
IFNA14 |
interferon, alpha 14 |
| chr12_-_118797475 | 0.03 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr19_-_22193731 | 0.03 |
ENST00000601773.1 ENST00000397126.4 ENST00000601993.1 ENST00000599916.1 |
ZNF208 |
zinc finger protein 208 |
| chr2_+_103089756 | 0.03 |
ENST00000295269.4 |
SLC9A4 |
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr22_-_32767017 | 0.02 |
ENST00000400234.1 |
RFPL3S |
RFPL3 antisense |
| chr14_-_36990354 | 0.02 |
ENST00000518149.1 |
NKX2-1 |
NK2 homeobox 1 |
| chr16_+_48278178 | 0.02 |
ENST00000285737.4 ENST00000535754.1 |
LONP2 |
lon peptidase 2, peroxisomal |
| chr17_+_1944790 | 0.02 |
ENST00000575162.1 |
DPH1 |
diphthamide biosynthesis 1 |
| chr11_-_13517565 | 0.02 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr17_-_72772462 | 0.02 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chr2_+_234826016 | 0.01 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr7_-_22862448 | 0.01 |
ENST00000358435.4 |
TOMM7 |
translocase of outer mitochondrial membrane 7 homolog (yeast) |
| chr11_-_4719072 | 0.01 |
ENST00000396950.3 ENST00000532598.1 |
OR51E2 |
olfactory receptor, family 51, subfamily E, member 2 |
| chr1_+_119957554 | 0.01 |
ENST00000543831.1 ENST00000433745.1 ENST00000369416.3 |
HSD3B2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr1_+_233086326 | 0.01 |
ENST00000366628.5 ENST00000366627.4 |
NTPCR |
nucleoside-triphosphatase, cancer-related |
| chr19_+_572543 | 0.01 |
ENST00000333511.3 ENST00000573216.1 ENST00000353555.4 |
BSG |
basigin (Ok blood group) |
| chr8_+_24151553 | 0.01 |
ENST00000265769.4 ENST00000540823.1 ENST00000397649.3 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr9_-_99540328 | 0.01 |
ENST00000223428.4 ENST00000375231.1 ENST00000374641.3 |
ZNF510 |
zinc finger protein 510 |
| chrX_-_154689596 | 0.01 |
ENST00000369444.2 |
H2AFB3 |
H2A histone family, member B3 |
| chr12_-_10282836 | 0.01 |
ENST00000304084.8 ENST00000353231.5 ENST00000525605.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr20_-_43589109 | 0.01 |
ENST00000372813.3 |
TOMM34 |
translocase of outer mitochondrial membrane 34 |
| chr11_-_89653576 | 0.01 |
ENST00000420869.1 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chr11_-_63376013 | 0.00 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chrX_+_154610428 | 0.00 |
ENST00000354514.4 |
H2AFB2 |
H2A histone family, member B2 |
| chr7_-_14026063 | 0.00 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr16_+_16434185 | 0.00 |
ENST00000524823.2 |
AC138969.4 |
Protein PKD1P1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.3 | 1.0 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 0.8 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.2 | 0.9 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.2 | 0.6 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.2 | 1.7 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.7 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 1.0 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.5 | GO:0034344 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.6 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 3.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.6 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.3 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.4 | GO:0046104 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 1.0 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.5 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.6 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.1 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.1 | 1.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.6 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.6 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 1.3 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 1.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 0.2 | GO:2000395 | ubiquitin-dependent endocytosis(GO:0070086) regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.9 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.4 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:1904502 | regulation of lipophagy(GO:1904502) positive regulation of lipophagy(GO:1904504) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.2 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.6 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.2 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.0 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.0 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.7 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.1 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.0 | 0.6 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 1.7 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.7 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.9 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.2 | 0.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.2 | 0.8 | GO:0047718 | enone reductase activity(GO:0035671) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.2 | 1.3 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.2 | 1.0 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.1 | 1.6 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 0.8 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.6 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 1.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.6 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 2.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 3.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.5 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 1.0 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.6 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.8 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.7 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.6 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 1.3 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 1.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.0 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 1.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |