Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
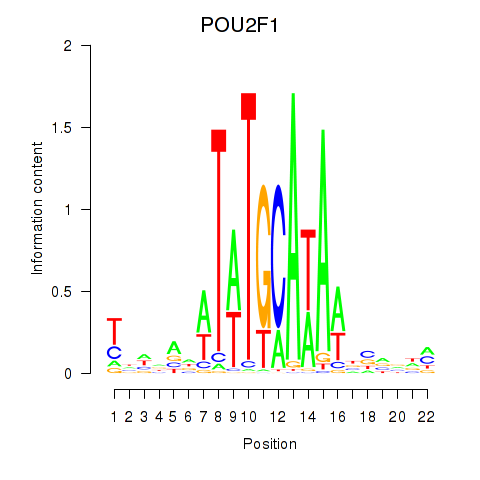
Results for POU2F1
Z-value: 0.57
Transcription factors associated with POU2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F1
|
ENSG00000143190.17 | POU2F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F1 | hg19_v2_chr1_+_167190066_167190156 | 0.56 | 2.4e-02 | Click! |
Activity profile of POU2F1 motif
Sorted Z-values of POU2F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU2F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_158801095 | 2.03 |
ENST00000368141.4 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr1_-_160681593 | 1.15 |
ENST00000368045.3 ENST00000368046.3 |
CD48 |
CD48 molecule |
| chr3_-_121379739 | 1.14 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr14_-_106733624 | 1.12 |
ENST00000390610.2 |
IGHV1-24 |
immunoglobulin heavy variable 1-24 |
| chr14_-_106471723 | 1.08 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr6_+_135502408 | 1.03 |
ENST00000341911.5 ENST00000442647.2 ENST00000316528.8 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr14_-_106539557 | 0.99 |
ENST00000390599.2 |
IGHV1-8 |
immunoglobulin heavy variable 1-8 |
| chr4_+_68424434 | 0.98 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr15_-_45670924 | 0.97 |
ENST00000396659.3 |
GATM |
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr6_+_135502466 | 0.97 |
ENST00000367814.4 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr19_-_9731872 | 0.93 |
ENST00000424629.1 ENST00000326044.5 ENST00000354661.4 ENST00000435550.1 ENST00000444611.1 ENST00000421525.1 |
ZNF561 |
zinc finger protein 561 |
| chr3_-_58200398 | 0.90 |
ENST00000318316.3 ENST00000460422.1 ENST00000483681.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr1_-_25291475 | 0.89 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr14_+_21249200 | 0.89 |
ENST00000304677.2 |
RNASE6 |
ribonuclease, RNase A family, k6 |
| chr15_-_22448819 | 0.84 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr12_-_102874416 | 0.61 |
ENST00000392904.1 ENST00000337514.6 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr14_-_106642049 | 0.59 |
ENST00000390605.2 |
IGHV1-18 |
immunoglobulin heavy variable 1-18 |
| chr15_-_55541227 | 0.59 |
ENST00000566877.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr12_-_102874378 | 0.53 |
ENST00000456098.1 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr8_+_86121448 | 0.51 |
ENST00000520225.1 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr14_-_107078851 | 0.50 |
ENST00000390628.2 |
IGHV1-58 |
immunoglobulin heavy variable 1-58 |
| chr21_+_10862622 | 0.50 |
ENST00000302092.5 ENST00000559480.1 |
IGHV1OR21-1 |
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr11_-_107729887 | 0.50 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr14_-_106453155 | 0.44 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr6_+_135502501 | 0.43 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr6_-_26216872 | 0.43 |
ENST00000244601.3 |
HIST1H2BG |
histone cluster 1, H2bg |
| chr2_-_89292422 | 0.39 |
ENST00000495489.1 |
IGKV1-8 |
immunoglobulin kappa variable 1-8 |
| chr18_-_52989217 | 0.39 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr4_+_106631966 | 0.36 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr2_-_89521942 | 0.34 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr7_+_7606497 | 0.34 |
ENST00000340080.4 ENST00000405785.1 ENST00000433635.1 |
MIOS |
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr2_+_89890533 | 0.34 |
ENST00000429992.2 |
IGKV2D-40 |
immunoglobulin kappa variable 2D-40 |
| chr16_+_19222479 | 0.33 |
ENST00000568433.1 |
SYT17 |
synaptotagmin XVII |
| chr9_+_116267536 | 0.32 |
ENST00000374136.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chrX_+_15518923 | 0.32 |
ENST00000348343.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr5_-_98262240 | 0.31 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr12_-_102874330 | 0.31 |
ENST00000307046.8 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chrX_-_135962876 | 0.31 |
ENST00000431446.3 ENST00000570135.1 ENST00000320676.7 ENST00000562646.1 |
RBMX |
RNA binding motif protein, X-linked |
| chr3_-_148939835 | 0.31 |
ENST00000264613.6 |
CP |
ceruloplasmin (ferroxidase) |
| chr16_+_33629600 | 0.30 |
ENST00000562905.2 |
IGHV3OR16-13 |
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr1_+_28562617 | 0.30 |
ENST00000497986.1 ENST00000335514.5 ENST00000468425.2 ENST00000465645.1 |
ATPIF1 |
ATPase inhibitory factor 1 |
| chr1_+_161736072 | 0.30 |
ENST00000367942.3 |
ATF6 |
activating transcription factor 6 |
| chr2_-_152684977 | 0.27 |
ENST00000428992.2 ENST00000295087.8 |
ARL5A |
ADP-ribosylation factor-like 5A |
| chr17_-_76573465 | 0.26 |
ENST00000585328.1 ENST00000389840.5 |
DNAH17 |
dynein, axonemal, heavy chain 17 |
| chr4_-_153274078 | 0.26 |
ENST00000263981.5 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr2_-_88285309 | 0.26 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chr15_+_48623208 | 0.25 |
ENST00000559935.1 ENST00000559416.1 |
DUT |
deoxyuridine triphosphatase |
| chr1_-_226926864 | 0.25 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr4_-_100356291 | 0.24 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr14_-_106967788 | 0.23 |
ENST00000390622.2 |
IGHV1-46 |
immunoglobulin heavy variable 1-46 |
| chr6_+_26217159 | 0.23 |
ENST00000303910.2 |
HIST1H2AE |
histone cluster 1, H2ae |
| chr5_+_129083772 | 0.23 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr8_-_33370607 | 0.23 |
ENST00000360742.5 ENST00000523305.1 |
TTI2 |
TELO2 interacting protein 2 |
| chr14_-_65346555 | 0.23 |
ENST00000542895.1 ENST00000556626.1 |
SPTB |
spectrin, beta, erythrocytic |
| chr7_+_28452130 | 0.22 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr7_+_64838786 | 0.22 |
ENST00000450302.2 |
ZNF92 |
zinc finger protein 92 |
| chr7_+_64838712 | 0.22 |
ENST00000328747.7 ENST00000431504.1 ENST00000357512.2 |
ZNF92 |
zinc finger protein 92 |
| chr13_+_111855414 | 0.21 |
ENST00000375737.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr2_-_74007193 | 0.20 |
ENST00000377706.4 ENST00000443070.1 ENST00000272444.3 |
DUSP11 |
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr6_+_122720681 | 0.18 |
ENST00000368455.4 ENST00000452194.1 |
HSF2 |
heat shock transcription factor 2 |
| chr14_+_65453432 | 0.18 |
ENST00000246166.2 |
FNTB |
farnesyltransferase, CAAX box, beta |
| chr3_-_48594248 | 0.18 |
ENST00000545984.1 ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr4_-_100356551 | 0.18 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr7_+_20686946 | 0.18 |
ENST00000443026.2 ENST00000406935.1 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr6_-_13621126 | 0.17 |
ENST00000600057.1 |
AL441883.1 |
Uncharacterized protein |
| chr12_-_110906027 | 0.17 |
ENST00000537466.2 ENST00000550974.1 ENST00000228827.3 |
GPN3 |
GPN-loop GTPase 3 |
| chr17_+_75181292 | 0.17 |
ENST00000431431.2 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
| chr5_+_67588391 | 0.17 |
ENST00000523872.1 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr15_+_43425672 | 0.16 |
ENST00000260403.2 |
TMEM62 |
transmembrane protein 62 |
| chr18_+_18943554 | 0.16 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr14_-_21270995 | 0.15 |
ENST00000555698.1 ENST00000397970.4 ENST00000340900.3 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr7_-_103848405 | 0.15 |
ENST00000447452.2 ENST00000545943.1 ENST00000297431.4 |
ORC5 |
origin recognition complex, subunit 5 |
| chr6_-_27782548 | 0.15 |
ENST00000333151.3 |
HIST1H2AJ |
histone cluster 1, H2aj |
| chr11_-_58343319 | 0.15 |
ENST00000395074.2 |
LPXN |
leupaxin |
| chr6_-_152489484 | 0.15 |
ENST00000354674.4 ENST00000539504.1 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr14_-_21270561 | 0.14 |
ENST00000412779.2 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr20_+_36373032 | 0.14 |
ENST00000373473.1 |
CTNNBL1 |
catenin, beta like 1 |
| chr3_-_46249878 | 0.13 |
ENST00000296140.3 |
CCR1 |
chemokine (C-C motif) receptor 1 |
| chr5_-_111093081 | 0.13 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr1_+_12977513 | 0.13 |
ENST00000330881.5 |
PRAMEF7 |
PRAME family member 7 |
| chr5_-_111092873 | 0.13 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr2_+_89999259 | 0.13 |
ENST00000558026.1 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr7_+_33765593 | 0.12 |
ENST00000311067.3 |
RP11-89N17.1 |
HCG1643653; Uncharacterized protein |
| chr9_-_21166659 | 0.12 |
ENST00000380225.1 |
IFNA21 |
interferon, alpha 21 |
| chr3_-_3221358 | 0.12 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr12_-_111358372 | 0.12 |
ENST00000548438.1 ENST00000228841.8 |
MYL2 |
myosin, light chain 2, regulatory, cardiac, slow |
| chr18_+_61144160 | 0.11 |
ENST00000489441.1 ENST00000424602.1 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr19_+_17858547 | 0.11 |
ENST00000600676.1 ENST00000600209.1 ENST00000596309.1 ENST00000598539.1 ENST00000597474.1 ENST00000593385.1 ENST00000598067.1 ENST00000593833.1 |
FCHO1 |
FCH domain only 1 |
| chr2_+_44001172 | 0.11 |
ENST00000260605.8 ENST00000406852.3 ENST00000443170.3 ENST00000398823.2 ENST00000605786.1 |
DYNC2LI1 |
dynein, cytoplasmic 2, light intermediate chain 1 |
| chr12_+_101988627 | 0.11 |
ENST00000547405.1 ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1 |
myosin binding protein C, slow type |
| chr8_-_117768023 | 0.11 |
ENST00000518949.1 ENST00000522453.1 ENST00000521861.1 ENST00000518995.1 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
| chr3_-_107777208 | 0.10 |
ENST00000398258.3 |
CD47 |
CD47 molecule |
| chr4_-_110723335 | 0.10 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr1_-_241683001 | 0.10 |
ENST00000366560.3 |
FH |
fumarate hydratase |
| chr1_-_223308098 | 0.10 |
ENST00000342210.6 |
TLR5 |
toll-like receptor 5 |
| chr16_-_18908196 | 0.09 |
ENST00000565324.1 ENST00000561947.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr14_-_107283278 | 0.09 |
ENST00000390639.2 |
IGHV7-81 |
immunoglobulin heavy variable 7-81 (non-functional) |
| chr20_+_39657454 | 0.09 |
ENST00000361337.2 |
TOP1 |
topoisomerase (DNA) I |
| chr2_+_103035102 | 0.09 |
ENST00000264260.2 |
IL18RAP |
interleukin 18 receptor accessory protein |
| chr4_-_144940477 | 0.08 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr9_-_35080013 | 0.08 |
ENST00000378643.3 |
FANCG |
Fanconi anemia, complementation group G |
| chr2_-_89459813 | 0.08 |
ENST00000390256.2 |
IGKV6-21 |
immunoglobulin kappa variable 6-21 (non-functional) |
| chr1_+_104293028 | 0.08 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr17_-_41623716 | 0.08 |
ENST00000319349.5 |
ETV4 |
ets variant 4 |
| chr4_-_110723194 | 0.07 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr10_+_95517566 | 0.07 |
ENST00000542308.1 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr21_-_33975547 | 0.07 |
ENST00000431599.1 |
C21orf59 |
chromosome 21 open reading frame 59 |
| chr6_+_26124373 | 0.07 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr3_-_33686743 | 0.07 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr6_-_26124138 | 0.07 |
ENST00000314332.5 ENST00000396984.1 |
HIST1H2BC |
histone cluster 1, H2bc |
| chr10_+_95517616 | 0.07 |
ENST00000371418.4 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr11_-_14521379 | 0.07 |
ENST00000249923.3 ENST00000529866.1 ENST00000439561.2 ENST00000534771.1 |
COPB1 |
coatomer protein complex, subunit beta 1 |
| chr15_-_55489097 | 0.06 |
ENST00000260443.4 |
RSL24D1 |
ribosomal L24 domain containing 1 |
| chrX_-_19988382 | 0.06 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr11_-_62609281 | 0.06 |
ENST00000525239.1 ENST00000538098.2 |
WDR74 |
WD repeat domain 74 |
| chr11_-_128457446 | 0.06 |
ENST00000392668.4 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr9_+_12693336 | 0.06 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr12_-_118797475 | 0.06 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr3_+_185046676 | 0.06 |
ENST00000428617.1 ENST00000443863.1 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chrX_-_130423386 | 0.06 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr10_+_79793518 | 0.05 |
ENST00000440692.1 ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24 |
ribosomal protein S24 |
| chr17_+_45728427 | 0.05 |
ENST00000540627.1 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr9_+_135854091 | 0.05 |
ENST00000450530.1 ENST00000534944.1 |
GFI1B |
growth factor independent 1B transcription repressor |
| chr1_+_104159999 | 0.05 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr9_-_123812542 | 0.05 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr3_-_167191814 | 0.05 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr10_-_115904361 | 0.05 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr3_+_196466710 | 0.05 |
ENST00000327134.3 |
PAK2 |
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr13_+_52436111 | 0.04 |
ENST00000242819.4 |
CCDC70 |
coiled-coil domain containing 70 |
| chr7_+_55433131 | 0.04 |
ENST00000254770.2 |
LANCL2 |
LanC lantibiotic synthetase component C-like 2 (bacterial) |
| chr9_-_86593238 | 0.04 |
ENST00000351839.3 |
HNRNPK |
heterogeneous nuclear ribonucleoprotein K |
| chr1_+_62439037 | 0.04 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr4_+_74702214 | 0.04 |
ENST00000226317.5 ENST00000515050.1 |
CXCL6 |
chemokine (C-X-C motif) ligand 6 |
| chr8_+_18067602 | 0.04 |
ENST00000307719.4 ENST00000545197.1 ENST00000539092.1 ENST00000541942.1 ENST00000518029.1 |
NAT1 |
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
| chr19_+_17858509 | 0.04 |
ENST00000594202.1 ENST00000252771.7 ENST00000389133.4 |
FCHO1 |
FCH domain only 1 |
| chr1_+_100436065 | 0.04 |
ENST00000370153.1 |
SLC35A3 |
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr22_+_41697520 | 0.04 |
ENST00000352645.4 |
ZC3H7B |
zinc finger CCCH-type containing 7B |
| chr22_-_29949680 | 0.03 |
ENST00000397873.2 ENST00000490103.1 |
THOC5 |
THO complex 5 |
| chr4_+_74301880 | 0.03 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr17_-_29624343 | 0.03 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chr10_+_95517660 | 0.03 |
ENST00000371413.3 |
LGI1 |
leucine-rich, glioma inactivated 1 |
| chr7_-_14026063 | 0.03 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr1_-_169703203 | 0.03 |
ENST00000333360.7 ENST00000367781.4 ENST00000367782.4 ENST00000367780.4 ENST00000367779.4 |
SELE |
selectin E |
| chr4_-_110723134 | 0.03 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr6_-_26043885 | 0.03 |
ENST00000357905.2 |
HIST1H2BB |
histone cluster 1, H2bb |
| chr17_-_64225508 | 0.02 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_+_90060377 | 0.02 |
ENST00000436451.2 |
IGKV6D-21 |
immunoglobulin kappa variable 6D-21 (non-functional) |
| chr1_-_109935819 | 0.02 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr14_-_68283291 | 0.02 |
ENST00000555452.1 ENST00000347230.4 |
ZFYVE26 |
zinc finger, FYVE domain containing 26 |
| chr6_-_46922659 | 0.01 |
ENST00000265417.7 |
GPR116 |
G protein-coupled receptor 116 |
| chrX_-_130423200 | 0.01 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr1_+_113010056 | 0.01 |
ENST00000369686.5 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
| chr2_+_204571198 | 0.01 |
ENST00000374481.3 ENST00000458610.2 ENST00000324106.8 |
CD28 |
CD28 molecule |
| chrX_-_130423240 | 0.01 |
ENST00000370910.1 ENST00000370901.4 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr6_+_72926145 | 0.01 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr10_+_13628933 | 0.01 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chrX_+_119384607 | 0.00 |
ENST00000326624.2 ENST00000557385.1 |
ZBTB33 |
zinc finger and BTB domain containing 33 |
| chr5_-_146781153 | 0.00 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr11_-_101000445 | 0.00 |
ENST00000534013.1 |
PGR |
progesterone receptor |
| chr16_+_14980632 | 0.00 |
ENST00000565655.1 |
NOMO1 |
NODAL modulator 1 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 4.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.2 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 2.0 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 2.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 1.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 1.6 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 1.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.3 | 1.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.3 | 1.4 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.4 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.6 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.3 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.1 | 2.0 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 1.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 4.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.2 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.3 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.9 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.2 | GO:0033029 | regulation of neutrophil apoptotic process(GO:0033029) common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0034146 | toll-like receptor 5 signaling pathway(GO:0034146) |
| 0.0 | 0.9 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.1 | GO:0050859 | negative regulation of B cell receptor signaling pathway(GO:0050859) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.3 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.9 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.3 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.2 | GO:1900103 | positive regulation of endoplasmic reticulum unfolded protein response(GO:1900103) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.2 | 1.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.4 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.1 | 0.3 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.1 | 0.3 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.1 | 4.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.3 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 1.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 1.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.3 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.2 | GO:0039552 | RIG-I binding(GO:0039552) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.9 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.9 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |