Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
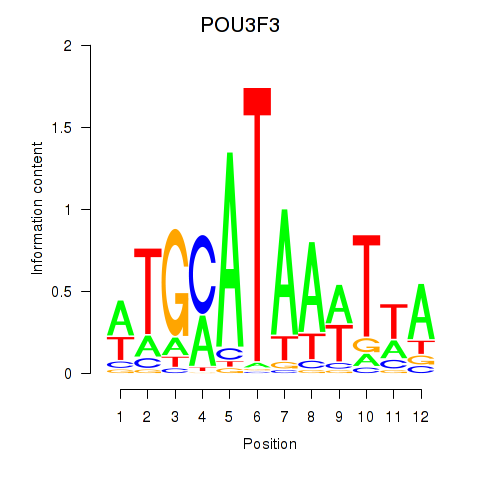
Results for POU3F3_POU3F4
Z-value: 1.29
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU3F3 |
|
POU3F4
|
ENSG00000196767.4 | POU3F4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU3F3 | hg19_v2_chr2_+_105471969_105471969 | 0.28 | 3.0e-01 | Click! |
| POU3F4 | hg19_v2_chrX_+_82763265_82763283 | -0.16 | 5.6e-01 | Click! |
Activity profile of POU3F3_POU3F4 motif
Sorted Z-values of POU3F3_POU3F4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU3F3_POU3F4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_189839046 | 4.71 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr8_-_49833978 | 3.46 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr10_-_75226166 | 3.40 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_125758813 | 3.19 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr12_-_91505608 | 3.15 |
ENST00000266718.4 |
LUM |
lumican |
| chr5_+_125758865 | 3.14 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chrX_+_65382433 | 3.03 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr5_+_119867159 | 3.00 |
ENST00000505123.1 |
PRR16 |
proline rich 16 |
| chr8_-_49834299 | 2.87 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr1_-_193155729 | 2.79 |
ENST00000367434.4 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr6_-_46459675 | 2.62 |
ENST00000306764.7 |
RCAN2 |
regulator of calcineurin 2 |
| chr13_+_102142296 | 2.10 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_-_91573249 | 2.07 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr3_+_158787041 | 2.04 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr3_+_35722487 | 1.96 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr3_-_107777208 | 1.87 |
ENST00000398258.3 |
CD47 |
CD47 molecule |
| chr11_-_2906979 | 1.82 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr11_+_2405833 | 1.73 |
ENST00000527343.1 ENST00000464784.2 |
CD81 |
CD81 molecule |
| chr5_+_140762268 | 1.68 |
ENST00000518325.1 |
PCDHGA7 |
protocadherin gamma subfamily A, 7 |
| chr18_-_52989217 | 1.67 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr13_+_102104980 | 1.51 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr17_-_10452929 | 1.48 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr14_-_61116168 | 1.42 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr2_-_175711133 | 1.42 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr18_-_52989525 | 1.40 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr19_-_44174330 | 1.37 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr12_-_91573316 | 1.37 |
ENST00000393155.1 |
DCN |
decorin |
| chr12_-_91574142 | 1.32 |
ENST00000547937.1 |
DCN |
decorin |
| chr12_-_91573132 | 1.31 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr5_-_133510456 | 1.30 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr12_-_50616382 | 1.17 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr1_-_1297157 | 1.16 |
ENST00000477278.2 |
MXRA8 |
matrix-remodelling associated 8 |
| chr19_-_44174305 | 1.10 |
ENST00000601723.1 ENST00000339082.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr5_+_140753444 | 1.04 |
ENST00000517434.1 |
PCDHGA6 |
protocadherin gamma subfamily A, 6 |
| chr3_-_127455200 | 0.94 |
ENST00000398101.3 |
MGLL |
monoglyceride lipase |
| chr8_-_27695552 | 0.94 |
ENST00000522944.1 ENST00000301905.4 |
PBK |
PDZ binding kinase |
| chr8_-_91095099 | 0.89 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr1_-_151345159 | 0.83 |
ENST00000458566.1 ENST00000447402.3 ENST00000426705.2 ENST00000435071.1 ENST00000368868.5 |
SELENBP1 |
selenium binding protein 1 |
| chr15_+_63569785 | 0.81 |
ENST00000380343.4 ENST00000560353.1 |
APH1B |
APH1B gamma secretase subunit |
| chr17_-_66951474 | 0.80 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr14_+_101297740 | 0.79 |
ENST00000555928.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr7_-_38948774 | 0.77 |
ENST00000395969.2 ENST00000414632.1 ENST00000310301.4 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr9_-_14180778 | 0.72 |
ENST00000380924.1 ENST00000543693.1 |
NFIB |
nuclear factor I/B |
| chr2_+_46769798 | 0.72 |
ENST00000238738.4 |
RHOQ |
ras homolog family member Q |
| chr10_-_49813090 | 0.72 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr17_-_3599327 | 0.70 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr3_-_141747950 | 0.70 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_3599696 | 0.68 |
ENST00000225328.5 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr5_+_141346385 | 0.68 |
ENST00000513019.1 ENST00000356143.1 |
RNF14 |
ring finger protein 14 |
| chr7_+_116502605 | 0.67 |
ENST00000458284.2 ENST00000490693.1 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr16_-_70835034 | 0.66 |
ENST00000261776.5 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
| chr1_+_59775752 | 0.66 |
ENST00000371212.1 |
FGGY |
FGGY carbohydrate kinase domain containing |
| chr15_+_63569731 | 0.66 |
ENST00000261879.5 |
APH1B |
APH1B gamma secretase subunit |
| chrX_+_80457442 | 0.65 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr6_+_26273144 | 0.65 |
ENST00000377733.2 |
HIST1H2BI |
histone cluster 1, H2bi |
| chr5_+_36152091 | 0.62 |
ENST00000274254.5 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr10_-_49812997 | 0.62 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr5_+_34757309 | 0.61 |
ENST00000397449.1 |
RAI14 |
retinoic acid induced 14 |
| chr3_-_141747439 | 0.61 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr5_+_36608422 | 0.61 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr5_+_36152179 | 0.55 |
ENST00000508514.1 ENST00000513151.1 ENST00000546211.1 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr2_+_161993465 | 0.52 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr10_-_90712520 | 0.51 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chrX_+_47053208 | 0.51 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr9_-_95244781 | 0.50 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr5_+_36152163 | 0.49 |
ENST00000274255.6 |
SKP2 |
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chrX_+_78003204 | 0.49 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr8_-_38008783 | 0.48 |
ENST00000276449.4 |
STAR |
steroidogenic acute regulatory protein |
| chr15_-_45670924 | 0.48 |
ENST00000396659.3 |
GATM |
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr3_-_196911002 | 0.47 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr5_+_159436120 | 0.46 |
ENST00000522793.1 ENST00000231238.5 |
TTC1 |
tetratricopeptide repeat domain 1 |
| chr12_-_71148413 | 0.46 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr12_-_71148357 | 0.45 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr2_-_55237484 | 0.44 |
ENST00000394609.2 |
RTN4 |
reticulon 4 |
| chr6_-_49712123 | 0.43 |
ENST00000263045.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr3_-_141747459 | 0.43 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_6614565 | 0.43 |
ENST00000377705.5 |
NOL9 |
nucleolar protein 9 |
| chr12_+_64173888 | 0.42 |
ENST00000537373.1 |
TMEM5 |
transmembrane protein 5 |
| chr8_-_120685608 | 0.41 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr4_-_144940477 | 0.40 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr5_-_111312622 | 0.40 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr2_+_33359687 | 0.40 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr2_+_33359646 | 0.40 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr5_+_140710061 | 0.40 |
ENST00000517417.1 ENST00000378105.3 |
PCDHGA1 |
protocadherin gamma subfamily A, 1 |
| chrX_-_119445306 | 0.37 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr2_+_161993412 | 0.35 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr6_-_49712147 | 0.34 |
ENST00000433368.2 ENST00000354620.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr2_+_239335449 | 0.34 |
ENST00000264607.4 |
ASB1 |
ankyrin repeat and SOCS box containing 1 |
| chr7_+_134528635 | 0.33 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr7_+_77469439 | 0.33 |
ENST00000450574.1 ENST00000416283.2 ENST00000248550.7 |
PHTF2 |
putative homeodomain transcription factor 2 |
| chr1_+_152627927 | 0.32 |
ENST00000444515.1 ENST00000536536.1 |
LINC00302 |
long intergenic non-protein coding RNA 302 |
| chr19_+_39138320 | 0.32 |
ENST00000424234.2 ENST00000390009.3 ENST00000589528.1 |
ACTN4 |
actinin, alpha 4 |
| chr17_-_38978847 | 0.32 |
ENST00000269576.5 |
KRT10 |
keratin 10 |
| chr15_+_59279851 | 0.31 |
ENST00000348370.4 ENST00000434298.1 ENST00000559160.1 |
RNF111 |
ring finger protein 111 |
| chr9_+_137967366 | 0.30 |
ENST00000252854.4 |
OLFM1 |
olfactomedin 1 |
| chr20_-_7921090 | 0.29 |
ENST00000378789.3 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr7_+_116502527 | 0.28 |
ENST00000361183.3 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr17_-_26220366 | 0.28 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr1_-_179112189 | 0.27 |
ENST00000512653.1 ENST00000344730.3 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr2_-_183387430 | 0.27 |
ENST00000410103.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr19_-_44123734 | 0.27 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr14_-_80697396 | 0.27 |
ENST00000557010.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr17_+_34171081 | 0.27 |
ENST00000585577.1 |
TAF15 |
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa |
| chr4_-_70826725 | 0.27 |
ENST00000353151.3 |
CSN2 |
casein beta |
| chr14_+_90722839 | 0.26 |
ENST00000261303.8 ENST00000553835.1 |
PSMC1 |
proteasome (prosome, macropain) 26S subunit, ATPase, 1 |
| chr21_-_27423339 | 0.26 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr7_+_150065278 | 0.25 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr14_+_90722886 | 0.25 |
ENST00000543772.2 |
PSMC1 |
proteasome (prosome, macropain) 26S subunit, ATPase, 1 |
| chr15_+_64680003 | 0.24 |
ENST00000261884.3 |
TRIP4 |
thyroid hormone receptor interactor 4 |
| chr12_+_59989918 | 0.24 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr15_+_66797627 | 0.24 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr4_-_144826682 | 0.24 |
ENST00000358615.4 ENST00000437468.2 |
GYPE |
glycophorin E (MNS blood group) |
| chrX_-_124097620 | 0.24 |
ENST00000371130.3 ENST00000422452.2 |
TENM1 |
teneurin transmembrane protein 1 |
| chr1_+_104159999 | 0.24 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr2_+_89999259 | 0.24 |
ENST00000558026.1 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr19_-_44124019 | 0.24 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr3_-_168865522 | 0.23 |
ENST00000464456.1 |
MECOM |
MDS1 and EVI1 complex locus |
| chr3_+_111805182 | 0.23 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chr6_-_131321863 | 0.23 |
ENST00000528282.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chr13_-_26795840 | 0.23 |
ENST00000381570.3 ENST00000399762.2 ENST00000346166.3 |
RNF6 |
ring finger protein (C3H2C3 type) 6 |
| chr8_-_93029865 | 0.23 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr22_-_39268308 | 0.23 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr8_-_41166953 | 0.23 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr5_+_141488070 | 0.22 |
ENST00000253814.4 |
NDFIP1 |
Nedd4 family interacting protein 1 |
| chr1_-_100231349 | 0.22 |
ENST00000287474.5 ENST00000414213.1 |
FRRS1 |
ferric-chelate reductase 1 |
| chr8_+_31496809 | 0.22 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr3_+_12525931 | 0.21 |
ENST00000446004.1 ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2 |
TSEN2 tRNA splicing endonuclease subunit |
| chr15_+_49447947 | 0.21 |
ENST00000327171.3 ENST00000560654.1 |
GALK2 |
galactokinase 2 |
| chr11_-_95657231 | 0.20 |
ENST00000409459.1 ENST00000352297.7 ENST00000393223.3 ENST00000346299.5 |
MTMR2 |
myotubularin related protein 2 |
| chr5_+_140529630 | 0.20 |
ENST00000543635.1 |
PCDHB6 |
protocadherin beta 6 |
| chrX_+_23682379 | 0.20 |
ENST00000379349.1 |
PRDX4 |
peroxiredoxin 4 |
| chr5_+_101569696 | 0.20 |
ENST00000597120.1 |
AC008948.1 |
AC008948.1 |
| chr4_-_168155730 | 0.19 |
ENST00000502330.1 ENST00000357154.3 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr1_-_62190793 | 0.19 |
ENST00000371177.2 ENST00000606498.1 |
TM2D1 |
TM2 domain containing 1 |
| chr1_+_101702417 | 0.19 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr1_-_179112173 | 0.18 |
ENST00000408940.3 ENST00000504405.1 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr11_+_33279850 | 0.17 |
ENST00000531504.1 ENST00000456517.1 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr15_-_30113676 | 0.17 |
ENST00000400011.2 |
TJP1 |
tight junction protein 1 |
| chr4_+_88754113 | 0.17 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr8_-_17942432 | 0.17 |
ENST00000381733.4 ENST00000314146.10 |
ASAH1 |
N-acylsphingosine amidohydrolase (acid ceramidase) 1 |
| chr7_+_80267973 | 0.17 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chrX_+_134975858 | 0.16 |
ENST00000537770.1 |
SAGE1 |
sarcoma antigen 1 |
| chr12_+_60058458 | 0.16 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr2_-_101034070 | 0.16 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr3_+_121902511 | 0.16 |
ENST00000490131.1 |
CASR |
calcium-sensing receptor |
| chr11_+_17316870 | 0.16 |
ENST00000458064.2 |
NUCB2 |
nucleobindin 2 |
| chr6_-_52859046 | 0.16 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr2_+_185463093 | 0.16 |
ENST00000302277.6 |
ZNF804A |
zinc finger protein 804A |
| chr4_+_186990298 | 0.16 |
ENST00000296795.3 ENST00000513189.1 |
TLR3 |
toll-like receptor 3 |
| chrX_+_134975753 | 0.16 |
ENST00000535938.1 |
SAGE1 |
sarcoma antigen 1 |
| chrX_+_54834004 | 0.16 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chrX_-_112084043 | 0.16 |
ENST00000304758.1 |
AMOT |
angiomotin |
| chr1_-_48866517 | 0.15 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr15_-_90892669 | 0.14 |
ENST00000412799.2 |
GABARAPL3 |
GABA(A) receptors associated protein like 3, pseudogene |
| chr8_+_31497271 | 0.14 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr14_+_70918874 | 0.14 |
ENST00000603540.1 |
ADAM21 |
ADAM metallopeptidase domain 21 |
| chrX_-_80457385 | 0.13 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr12_+_110011571 | 0.13 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr12_-_111926342 | 0.13 |
ENST00000389154.3 |
ATXN2 |
ataxin 2 |
| chr14_-_25479811 | 0.13 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr1_-_109935819 | 0.13 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr12_+_27175476 | 0.12 |
ENST00000546323.1 ENST00000282892.3 |
MED21 |
mediator complex subunit 21 |
| chr10_+_35484793 | 0.12 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr22_-_43036607 | 0.12 |
ENST00000505920.1 |
ATP5L2 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G2 |
| chr1_+_24019099 | 0.11 |
ENST00000443624.1 ENST00000458455.1 |
RPL11 |
ribosomal protein L11 |
| chr1_+_212606219 | 0.11 |
ENST00000366988.3 |
NENF |
neudesin neurotrophic factor |
| chr12_-_10324716 | 0.11 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr17_-_39023462 | 0.11 |
ENST00000251643.4 |
KRT12 |
keratin 12 |
| chr12_-_102224457 | 0.11 |
ENST00000549165.1 ENST00000549940.1 ENST00000392919.4 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr19_+_39138271 | 0.10 |
ENST00000252699.2 |
ACTN4 |
actinin, alpha 4 |
| chr15_-_74658493 | 0.10 |
ENST00000419019.2 ENST00000569662.1 |
CYP11A1 |
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr12_-_7656357 | 0.10 |
ENST00000396620.3 ENST00000432237.2 ENST00000359156.4 |
CD163 |
CD163 molecule |
| chr9_+_77230499 | 0.10 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr3_-_52931557 | 0.10 |
ENST00000504329.1 ENST00000355083.5 |
TMEM110-MUSTN1 TMEM110 |
TMEM110-MUSTN1 readthrough transmembrane protein 110 |
| chr5_+_140782351 | 0.10 |
ENST00000573521.1 |
PCDHGA9 |
protocadherin gamma subfamily A, 9 |
| chrX_+_65382381 | 0.09 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr2_-_183387283 | 0.09 |
ENST00000435564.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr13_-_46716969 | 0.09 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_+_130569429 | 0.09 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_104293028 | 0.08 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr4_+_70796784 | 0.08 |
ENST00000246891.4 ENST00000444405.3 |
CSN1S1 |
casein alpha s1 |
| chr17_+_45608430 | 0.08 |
ENST00000322157.4 |
NPEPPS |
aminopeptidase puromycin sensitive |
| chr2_+_201170703 | 0.08 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr9_+_137967268 | 0.08 |
ENST00000371799.4 ENST00000277415.11 |
OLFM1 |
olfactomedin 1 |
| chr4_+_175839551 | 0.07 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr11_-_62369291 | 0.07 |
ENST00000278823.2 |
MTA2 |
metastasis associated 1 family, member 2 |
| chr7_+_106505912 | 0.07 |
ENST00000359195.3 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr6_+_29429217 | 0.07 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr15_+_74422585 | 0.07 |
ENST00000561740.1 ENST00000435464.1 ENST00000453268.2 |
ISLR2 |
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr11_-_28129656 | 0.07 |
ENST00000263181.6 |
KIF18A |
kinesin family member 18A |
| chr22_+_40322623 | 0.06 |
ENST00000399090.2 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr6_-_32374900 | 0.06 |
ENST00000374995.3 ENST00000374993.1 ENST00000414363.1 ENST00000540315.1 ENST00000544175.1 ENST00000429232.2 ENST00000454136.3 ENST00000446536.2 |
BTNL2 |
butyrophilin-like 2 (MHC class II associated) |
| chr22_+_40322595 | 0.06 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr17_-_61996136 | 0.06 |
ENST00000342364.4 |
GH1 |
growth hormone 1 |
| chr15_+_71228826 | 0.06 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr5_-_160973649 | 0.06 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr8_-_59412717 | 0.06 |
ENST00000301645.3 |
CYP7A1 |
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr9_+_34458771 | 0.06 |
ENST00000437363.1 ENST00000242317.4 |
DNAI1 |
dynein, axonemal, intermediate chain 1 |
| chr3_-_151047327 | 0.06 |
ENST00000325602.5 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
| chr2_-_220264703 | 0.06 |
ENST00000519905.1 ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP |
aspartyl aminopeptidase |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.6 | 2.5 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.6 | 4.7 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.5 | 6.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.5 | 1.4 | GO:0071698 | olfactory placode formation(GO:0030910) myotome development(GO:0061055) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.4 | 1.7 | GO:0043128 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.4 | 3.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.3 | 1.7 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.3 | 2.8 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.3 | 3.4 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.3 | 1.8 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 3.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 0.9 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.2 | 0.5 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.2 | 0.8 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.9 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 1.3 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 3.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 1.5 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.1 | 1.5 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.7 | GO:2000795 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.9 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.2 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.1 | 0.3 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.3 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.1 | 0.3 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.8 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.6 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.5 | GO:0090131 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) mesenchyme migration(GO:0090131) |
| 0.1 | 1.5 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.2 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 0.8 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.2 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.1 | 0.5 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.1 | 0.3 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 2.6 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 | 0.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.2 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.4 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.5 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 1.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:2000255 | regulation of male germ cell proliferation(GO:2000254) negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 1.4 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0034343 | microglial cell activation involved in immune response(GO:0002282) type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 1.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.7 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 2.1 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.3 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.0 | GO:0090264 | immune complex clearance(GO:0002434) immune complex clearance by monocytes and macrophages(GO:0002436) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) |
| 0.0 | 1.3 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 3.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.7 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:1904778 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.0 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.3 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.6 | GO:0009967 | positive regulation of signal transduction(GO:0009967) |
| 0.0 | 0.3 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 3.1 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.1 | GO:0002091 | negative regulation of receptor internalization(GO:0002091) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.9 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.2 | GO:0006091 | generation of precursor metabolites and energy(GO:0006091) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 9.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.5 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 5.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 4.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.7 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 3.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.7 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.8 | 2.5 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.4 | 2.6 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 4.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.3 | 3.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 3.3 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.7 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.2 | 3.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.2 | 2.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.2 | 0.8 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.9 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 0.9 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 1.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.1 | 1.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 9.7 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.8 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.4 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.3 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 1.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 6.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 1.5 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.9 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.3 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.5 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.4 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.5 | 1.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.4 | 6.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.4 | 3.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 2.5 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.2 | 0.8 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 1.3 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 1.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.5 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.7 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 0.5 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 1.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 1.0 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.7 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.1 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.5 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.8 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.7 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.7 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.0 | 5.0 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.4 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0044438 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.1 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.0 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 1.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 2.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 4.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 3.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 2.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 1.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.8 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |