Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
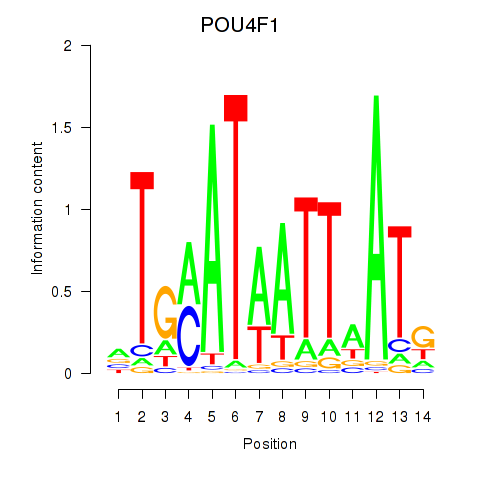
Results for POU4F1_POU4F3
Z-value: 0.50
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F1
|
ENSG00000152192.6 | POU4F1 |
|
POU4F3
|
ENSG00000091010.4 | POU4F3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F1 | hg19_v2_chr13_-_79177673_79177701 | -0.20 | 4.5e-01 | Click! |
| POU4F3 | hg19_v2_chr5_+_145718587_145718607 | -0.00 | 9.9e-01 | Click! |
Activity profile of POU4F1_POU4F3 motif
Sorted Z-values of POU4F1_POU4F3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F1_POU4F3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_65409438 | 1.71 |
ENST00000557049.1 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr4_+_74269956 | 1.58 |
ENST00000295897.4 ENST00000415165.2 ENST00000503124.1 ENST00000509063.1 ENST00000401494.3 |
ALB |
albumin |
| chr14_-_65409502 | 1.50 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr17_-_56492989 | 0.83 |
ENST00000583753.1 |
RNF43 |
ring finger protein 43 |
| chr12_+_49740700 | 0.78 |
ENST00000549441.2 ENST00000395069.3 |
DNAJC22 |
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr1_+_66458072 | 0.77 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr4_-_16085314 | 0.77 |
ENST00000510224.1 |
PROM1 |
prominin 1 |
| chr4_-_16085340 | 0.73 |
ENST00000508167.1 |
PROM1 |
prominin 1 |
| chr9_-_123812542 | 0.71 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr5_+_129083772 | 0.62 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr7_-_22234381 | 0.57 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr19_-_36304201 | 0.57 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr15_+_58724184 | 0.56 |
ENST00000433326.2 |
LIPC |
lipase, hepatic |
| chr15_-_20193370 | 0.53 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr11_+_22696314 | 0.52 |
ENST00000532398.1 ENST00000433790.1 |
GAS2 |
growth arrest-specific 2 |
| chr15_-_52587945 | 0.49 |
ENST00000443683.2 ENST00000558479.1 ENST00000261839.7 |
MYO5C |
myosin VC |
| chr17_-_40540586 | 0.48 |
ENST00000264657.5 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr22_+_40322595 | 0.43 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr22_+_40322623 | 0.42 |
ENST00000399090.2 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr10_-_11574274 | 0.42 |
ENST00000277575.5 |
USP6NL |
USP6 N-terminal like |
| chr17_-_40540377 | 0.40 |
ENST00000404395.3 ENST00000389272.3 ENST00000585517.1 ENST00000588065.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr3_-_149375783 | 0.40 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr3_-_194072019 | 0.38 |
ENST00000429275.1 ENST00000323830.3 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
| chrX_-_32173579 | 0.36 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr17_-_40540484 | 0.34 |
ENST00000588969.1 |
STAT3 |
signal transducer and activator of transcription 3 (acute-phase response factor) |
| chr8_+_11666649 | 0.33 |
ENST00000528643.1 ENST00000525777.1 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chrX_-_15288154 | 0.33 |
ENST00000380483.3 ENST00000380485.3 ENST00000380488.4 |
ASB9 |
ankyrin repeat and SOCS box containing 9 |
| chr2_-_88285309 | 0.33 |
ENST00000420840.2 |
RGPD2 |
RANBP2-like and GRIP domain containing 2 |
| chr12_-_6798616 | 0.32 |
ENST00000355772.4 ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384 |
zinc finger protein 384 |
| chr17_+_7155819 | 0.31 |
ENST00000570322.1 ENST00000576496.1 ENST00000574841.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr18_-_31803435 | 0.30 |
ENST00000589544.1 ENST00000269185.4 ENST00000261592.5 |
NOL4 |
nucleolar protein 4 |
| chr12_-_6798410 | 0.29 |
ENST00000361959.3 ENST00000436774.2 ENST00000544482.1 |
ZNF384 |
zinc finger protein 384 |
| chr12_-_6798523 | 0.28 |
ENST00000319770.3 |
ZNF384 |
zinc finger protein 384 |
| chr2_+_170590321 | 0.27 |
ENST00000392647.2 |
KLHL23 |
kelch-like family member 23 |
| chr22_-_39268308 | 0.26 |
ENST00000407418.3 |
CBX6 |
chromobox homolog 6 |
| chr1_+_73771844 | 0.25 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr1_+_50574585 | 0.24 |
ENST00000371824.1 ENST00000371823.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_100026406 | 0.24 |
ENST00000414441.1 |
MEPCE |
methylphosphate capping enzyme |
| chrX_-_39956656 | 0.23 |
ENST00000397354.3 ENST00000378444.4 |
BCOR |
BCL6 corepressor |
| chr14_+_21249200 | 0.23 |
ENST00000304677.2 |
RNASE6 |
ribonuclease, RNase A family, k6 |
| chr1_-_160549235 | 0.22 |
ENST00000368054.3 ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84 |
CD84 molecule |
| chr4_-_69817481 | 0.22 |
ENST00000251566.4 |
UGT2A3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr16_-_51185172 | 0.21 |
ENST00000251020.4 |
SALL1 |
spalt-like transcription factor 1 |
| chr20_+_36405665 | 0.21 |
ENST00000373469.1 |
CTNNBL1 |
catenin, beta like 1 |
| chr4_+_68424434 | 0.21 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chrX_+_47053208 | 0.20 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr1_+_198608146 | 0.20 |
ENST00000367376.2 ENST00000352140.3 ENST00000594404.1 ENST00000598951.1 ENST00000530727.1 ENST00000442510.2 ENST00000367367.4 ENST00000348564.6 ENST00000367364.1 ENST00000413409.2 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
| chrX_-_138724994 | 0.19 |
ENST00000536274.1 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr14_-_107049312 | 0.19 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chr18_-_31803169 | 0.19 |
ENST00000590712.1 |
NOL4 |
nucleolar protein 4 |
| chr14_-_106668095 | 0.19 |
ENST00000390606.2 |
IGHV3-20 |
immunoglobulin heavy variable 3-20 |
| chr17_-_29641084 | 0.18 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr12_+_861717 | 0.18 |
ENST00000535572.1 |
WNK1 |
WNK lysine deficient protein kinase 1 |
| chr3_-_185826855 | 0.17 |
ENST00000306376.5 |
ETV5 |
ets variant 5 |
| chr2_-_101034070 | 0.17 |
ENST00000264249.3 |
CHST10 |
carbohydrate sulfotransferase 10 |
| chr11_-_35441524 | 0.17 |
ENST00000395750.1 ENST00000449068.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_-_183387064 | 0.16 |
ENST00000536095.1 ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr2_-_183106641 | 0.15 |
ENST00000346717.4 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr15_+_25068773 | 0.14 |
ENST00000400100.1 ENST00000400098.1 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr10_+_135207623 | 0.14 |
ENST00000317502.6 ENST00000432508.3 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr3_-_149470229 | 0.14 |
ENST00000473414.1 |
COMMD2 |
COMM domain containing 2 |
| chr12_-_42631529 | 0.14 |
ENST00000548917.1 |
YAF2 |
YY1 associated factor 2 |
| chr18_+_47088401 | 0.14 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr19_+_54466179 | 0.13 |
ENST00000270458.2 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
| chr9_+_35673853 | 0.13 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr2_-_183387283 | 0.13 |
ENST00000435564.1 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
| chr10_+_135207598 | 0.13 |
ENST00000477902.2 |
MTG1 |
mitochondrial ribosome-associated GTPase 1 |
| chr4_+_106631966 | 0.13 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr15_-_65426174 | 0.13 |
ENST00000204549.4 |
PDCD7 |
programmed cell death 7 |
| chr6_-_44281043 | 0.13 |
ENST00000244571.4 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
| chr12_-_89746173 | 0.13 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr13_-_46756351 | 0.12 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr14_-_21270995 | 0.12 |
ENST00000555698.1 ENST00000397970.4 ENST00000340900.3 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr22_-_30970560 | 0.12 |
ENST00000402369.1 ENST00000406361.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr1_+_81771806 | 0.12 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr7_-_20256965 | 0.12 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr1_+_101003687 | 0.12 |
ENST00000315033.4 |
GPR88 |
G protein-coupled receptor 88 |
| chr3_-_52860850 | 0.12 |
ENST00000441637.2 |
ITIH4 |
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr3_-_58613323 | 0.12 |
ENST00000474531.1 ENST00000465970.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr7_-_100026280 | 0.11 |
ENST00000360951.4 ENST00000398027.2 ENST00000324725.6 ENST00000472716.1 |
ZCWPW1 |
zinc finger, CW type with PWWP domain 1 |
| chrX_-_138724677 | 0.11 |
ENST00000370573.4 ENST00000338585.6 ENST00000370576.4 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr14_+_39703084 | 0.11 |
ENST00000553728.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr1_-_95538492 | 0.11 |
ENST00000370205.5 |
ALG14 |
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| chr12_-_23737534 | 0.11 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr11_-_8290263 | 0.11 |
ENST00000428101.2 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
| chr4_+_102711764 | 0.10 |
ENST00000322953.4 ENST00000428908.1 |
BANK1 |
B-cell scaffold protein with ankyrin repeats 1 |
| chr1_-_154580616 | 0.10 |
ENST00000368474.4 |
ADAR |
adenosine deaminase, RNA-specific |
| chr14_-_21270561 | 0.10 |
ENST00000412779.2 |
RNASE1 |
ribonuclease, RNase A family, 1 (pancreatic) |
| chr3_+_70048881 | 0.10 |
ENST00000483525.1 |
RP11-460N16.1 |
RP11-460N16.1 |
| chr2_+_87135076 | 0.09 |
ENST00000409776.2 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
| chr16_+_48278178 | 0.09 |
ENST00000285737.4 ENST00000535754.1 |
LONP2 |
lon peptidase 2, peroxisomal |
| chr22_+_39101728 | 0.09 |
ENST00000216044.5 ENST00000484657.1 |
GTPBP1 |
GTP binding protein 1 |
| chr7_-_115670792 | 0.09 |
ENST00000265440.7 ENST00000393485.1 |
TFEC |
transcription factor EC |
| chr5_-_11588907 | 0.09 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr14_+_39583427 | 0.09 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr12_-_100486668 | 0.09 |
ENST00000550544.1 ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L |
UHRF1 binding protein 1-like |
| chr6_-_76782371 | 0.09 |
ENST00000369950.3 ENST00000369963.3 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
| chr11_-_35441597 | 0.08 |
ENST00000395753.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_-_11589131 | 0.08 |
ENST00000511377.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr17_+_45728427 | 0.08 |
ENST00000540627.1 |
KPNB1 |
karyopherin (importin) beta 1 |
| chr8_-_125577940 | 0.08 |
ENST00000519168.1 ENST00000395508.2 |
MTSS1 |
metastasis suppressor 1 |
| chr1_+_67773044 | 0.08 |
ENST00000262345.1 ENST00000371000.1 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
| chr16_+_31366536 | 0.08 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr7_-_115670804 | 0.08 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr12_+_16109519 | 0.08 |
ENST00000526530.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr2_+_87144738 | 0.07 |
ENST00000559485.1 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
| chr14_-_54908043 | 0.07 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr6_+_126102292 | 0.07 |
ENST00000368357.3 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr14_+_31494672 | 0.07 |
ENST00000542754.2 ENST00000313566.6 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr7_-_100425112 | 0.07 |
ENST00000358173.3 |
EPHB4 |
EPH receptor B4 |
| chr9_-_21166659 | 0.06 |
ENST00000380225.1 |
IFNA21 |
interferon, alpha 21 |
| chr19_-_18632861 | 0.06 |
ENST00000262809.4 |
ELL |
elongation factor RNA polymerase II |
| chr10_-_115904361 | 0.06 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr4_-_122085469 | 0.06 |
ENST00000057513.3 |
TNIP3 |
TNFAIP3 interacting protein 3 |
| chrX_+_41583408 | 0.06 |
ENST00000302548.4 |
GPR82 |
G protein-coupled receptor 82 |
| chr20_+_57875758 | 0.06 |
ENST00000395654.3 |
EDN3 |
endothelin 3 |
| chr1_+_79115503 | 0.06 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr19_-_53696587 | 0.06 |
ENST00000396424.3 ENST00000600412.1 |
ZNF665 |
zinc finger protein 665 |
| chr1_-_54303934 | 0.06 |
ENST00000537333.1 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr4_-_122744998 | 0.06 |
ENST00000274026.5 |
CCNA2 |
cyclin A2 |
| chr19_+_16940198 | 0.05 |
ENST00000248054.5 ENST00000596802.1 ENST00000379803.1 |
SIN3B |
SIN3 transcription regulator family member B |
| chr1_+_160336851 | 0.05 |
ENST00000302101.5 |
NHLH1 |
nescient helix loop helix 1 |
| chr1_-_54303949 | 0.05 |
ENST00000234725.8 |
NDC1 |
NDC1 transmembrane nucleoporin |
| chr12_-_51611477 | 0.05 |
ENST00000389243.4 |
POU6F1 |
POU class 6 homeobox 1 |
| chr10_+_104613980 | 0.05 |
ENST00000339834.5 |
C10orf32 |
chromosome 10 open reading frame 32 |
| chr18_+_32397918 | 0.05 |
ENST00000590727.1 ENST00000601125.1 |
DTNA |
dystrobrevin, alpha |
| chr21_-_37914898 | 0.05 |
ENST00000399136.1 |
CLDN14 |
claudin 14 |
| chr5_-_134871639 | 0.05 |
ENST00000314744.4 |
NEUROG1 |
neurogenin 1 |
| chr17_+_29664830 | 0.04 |
ENST00000444181.2 ENST00000417592.2 |
NF1 |
neurofibromin 1 |
| chr5_+_140501581 | 0.04 |
ENST00000194152.1 |
PCDHB4 |
protocadherin beta 4 |
| chr13_-_70682590 | 0.04 |
ENST00000377844.4 |
KLHL1 |
kelch-like family member 1 |
| chr17_-_50236039 | 0.04 |
ENST00000451037.2 |
CA10 |
carbonic anhydrase X |
| chr16_+_31366455 | 0.04 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr1_+_158900568 | 0.04 |
ENST00000458222.1 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr11_+_120973375 | 0.04 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr5_-_77844974 | 0.04 |
ENST00000515007.2 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
| chr1_-_156470515 | 0.03 |
ENST00000340875.5 ENST00000368240.2 ENST00000353795.3 |
MEF2D |
myocyte enhancer factor 2D |
| chr5_-_41261540 | 0.03 |
ENST00000263413.3 |
C6 |
complement component 6 |
| chr16_+_619931 | 0.03 |
ENST00000321878.5 ENST00000439574.1 ENST00000026218.5 ENST00000470411.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr12_+_41831485 | 0.03 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr15_+_74422585 | 0.03 |
ENST00000561740.1 ENST00000435464.1 ENST00000453268.2 |
ISLR2 |
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr14_+_31494841 | 0.03 |
ENST00000556232.1 ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr18_+_61575200 | 0.03 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr4_+_37962018 | 0.03 |
ENST00000504686.1 |
PTTG2 |
pituitary tumor-transforming 2 |
| chr18_+_29027696 | 0.03 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr10_-_6104253 | 0.02 |
ENST00000256876.6 ENST00000379954.1 ENST00000379959.3 |
IL2RA |
interleukin 2 receptor, alpha |
| chr13_+_52586517 | 0.02 |
ENST00000523764.1 ENST00000521508.1 |
ALG11 |
ALG11, alpha-1,2-mannosyltransferase |
| chrX_-_107975917 | 0.02 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chr20_+_30063067 | 0.02 |
ENST00000201979.2 |
REM1 |
RAS (RAD and GEM)-like GTP-binding 1 |
| chr5_-_58295712 | 0.02 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr2_-_70475730 | 0.02 |
ENST00000445587.1 ENST00000433529.2 ENST00000415783.2 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr1_-_216978709 | 0.02 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr10_-_22292613 | 0.02 |
ENST00000376980.3 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr22_-_43010928 | 0.02 |
ENST00000348657.2 ENST00000252115.5 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
| chr17_+_18759612 | 0.01 |
ENST00000432893.2 ENST00000414602.1 ENST00000574522.1 ENST00000570450.1 ENST00000419071.2 |
PRPSAP2 |
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr9_+_135906076 | 0.01 |
ENST00000372097.5 ENST00000440319.1 |
GTF3C5 |
general transcription factor IIIC, polypeptide 5, 63kDa |
| chr12_-_102224704 | 0.01 |
ENST00000299314.7 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr4_-_68749699 | 0.01 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr2_+_204732487 | 0.01 |
ENST00000302823.3 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
| chr1_-_33430286 | 0.01 |
ENST00000373456.7 ENST00000356990.5 ENST00000235150.4 |
RNF19B |
ring finger protein 19B |
| chr4_-_25032501 | 0.01 |
ENST00000382114.4 |
LGI2 |
leucine-rich repeat LGI family, member 2 |
| chr9_-_215744 | 0.01 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr10_+_79793518 | 0.01 |
ENST00000440692.1 ENST00000435275.1 ENST00000372360.3 ENST00000360830.4 |
RPS24 |
ribosomal protein S24 |
| chr12_-_111926342 | 0.01 |
ENST00000389154.3 |
ATXN2 |
ataxin 2 |
| chr9_+_34458771 | 0.01 |
ENST00000437363.1 ENST00000242317.4 |
DNAI1 |
dynein, axonemal, intermediate chain 1 |
| chr2_+_202047843 | 0.00 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr2_+_29320571 | 0.00 |
ENST00000401605.1 ENST00000401617.2 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr11_+_86085778 | 0.00 |
ENST00000354755.1 ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81 |
coiled-coil domain containing 81 |
| chr12_+_10658201 | 0.00 |
ENST00000322446.3 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chrX_+_65382381 | 0.00 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr17_-_49124230 | 0.00 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 1.2 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 3.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 0.3 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.6 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 1.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.1 | GO:0000806 | Y chromosome(GO:0000806) |
| 0.0 | 0.4 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.4 | 1.5 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.3 | 1.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.3 | 0.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.2 | GO:0072092 | olfactory bulb mitral cell layer development(GO:0061034) ureteric bud invasion(GO:0072092) |
| 0.1 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.8 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.2 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.0 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.2 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0048539 | immunoglobulin biosynthetic process(GO:0002378) bone marrow development(GO:0048539) |
| 0.0 | 0.6 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.2 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.1 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.4 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 3.0 | GO:0098869 | cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.3 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.4 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 0.1 | GO:0098722 | asymmetric neuroblast division(GO:0055059) asymmetric stem cell division(GO:0098722) |
| 0.0 | 0.5 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.7 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 3.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |