Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
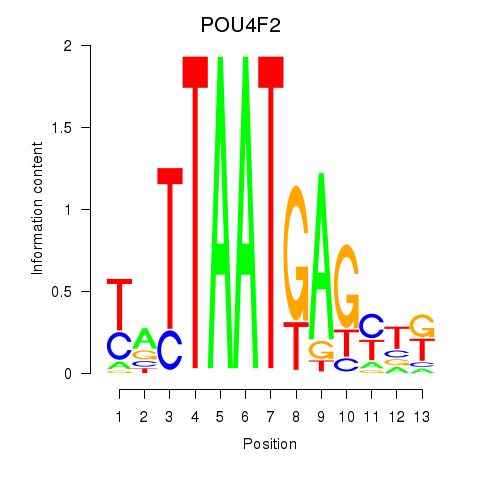
Results for POU4F2
Z-value: 0.50
Transcription factors associated with POU4F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F2
|
ENSG00000151615.3 | POU4F2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU4F2 | hg19_v2_chr4_+_147560042_147560046 | -0.33 | 2.1e-01 | Click! |
Activity profile of POU4F2 motif
Sorted Z-values of POU4F2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU4F2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_49834299 | 1.70 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr8_-_18541603 | 1.61 |
ENST00000428502.2 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr12_-_89746173 | 1.58 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr8_-_49833978 | 1.57 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr4_+_169418195 | 1.54 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr3_-_79816965 | 1.50 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr4_+_169418255 | 1.11 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr7_-_81399329 | 0.90 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr7_-_81399438 | 0.59 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr9_+_125132803 | 0.47 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr14_-_70883708 | 0.47 |
ENST00000256366.4 |
SYNJ2BP |
synaptojanin 2 binding protein |
| chr3_+_159557637 | 0.46 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr1_+_40506392 | 0.43 |
ENST00000414893.1 ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr7_-_81399411 | 0.42 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr17_-_64225508 | 0.41 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr9_+_125133315 | 0.35 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr7_-_81399355 | 0.32 |
ENST00000457544.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr6_-_109777128 | 0.31 |
ENST00000358807.3 ENST00000358577.3 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr5_-_134914673 | 0.27 |
ENST00000512158.1 |
CXCL14 |
chemokine (C-X-C motif) ligand 14 |
| chr13_-_36050819 | 0.27 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr12_-_30887948 | 0.23 |
ENST00000433722.2 |
CAPRIN2 |
caprin family member 2 |
| chr4_-_10686475 | 0.22 |
ENST00000226951.6 |
CLNK |
cytokine-dependent hematopoietic cell linker |
| chr18_-_71959159 | 0.20 |
ENST00000494131.2 ENST00000397914.4 ENST00000340533.4 |
CYB5A |
cytochrome b5 type A (microsomal) |
| chr6_+_39760129 | 0.18 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr12_+_109577202 | 0.15 |
ENST00000377848.3 ENST00000377854.5 |
ACACB |
acetyl-CoA carboxylase beta |
| chr18_+_28898052 | 0.15 |
ENST00000257192.4 |
DSG1 |
desmoglein 1 |
| chr17_-_7832753 | 0.14 |
ENST00000303790.2 |
KCNAB3 |
potassium voltage-gated channel, shaker-related subfamily, beta member 3 |
| chr14_+_32030582 | 0.14 |
ENST00000550649.1 ENST00000281081.7 |
NUBPL |
nucleotide binding protein-like |
| chr20_+_56136136 | 0.12 |
ENST00000319441.4 ENST00000543666.1 |
PCK1 |
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr12_-_118797475 | 0.12 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr19_+_1269324 | 0.11 |
ENST00000589710.1 ENST00000588230.1 ENST00000413636.2 ENST00000586472.1 ENST00000589686.1 ENST00000444172.2 ENST00000587323.1 ENST00000320936.5 ENST00000587896.1 ENST00000589235.1 ENST00000591659.1 |
CIRBP |
cold inducible RNA binding protein |
| chrX_-_18690210 | 0.11 |
ENST00000379984.3 |
RS1 |
retinoschisin 1 |
| chr4_+_169013666 | 0.11 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr1_+_229440129 | 0.09 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr5_-_140998481 | 0.09 |
ENST00000518047.1 |
DIAPH1 |
diaphanous-related formin 1 |
| chr1_-_44482979 | 0.08 |
ENST00000360584.2 ENST00000357730.2 ENST00000528803.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr8_+_22857048 | 0.08 |
ENST00000251822.6 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
| chr5_+_66300446 | 0.07 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_118796910 | 0.06 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr11_-_125550764 | 0.05 |
ENST00000527795.1 |
ACRV1 |
acrosomal vesicle protein 1 |
| chr6_-_111136513 | 0.05 |
ENST00000368911.3 |
CDK19 |
cyclin-dependent kinase 19 |
| chr11_+_118175132 | 0.04 |
ENST00000361763.4 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
| chr12_+_53848505 | 0.03 |
ENST00000552819.1 ENST00000455667.3 |
PCBP2 |
poly(rC) binding protein 2 |
| chr14_+_55221541 | 0.03 |
ENST00000555192.1 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chr2_+_44502597 | 0.03 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr2_-_70780572 | 0.01 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr12_+_56510368 | 0.01 |
ENST00000546591.1 ENST00000501597.3 |
RPL41 |
ribosomal protein L41 |
| chrX_+_68835911 | 0.00 |
ENST00000525810.1 ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA |
ectodysplasin A |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 1.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.5 | 1.5 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.4 | 1.6 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 2.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 2.6 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 1.6 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.8 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.2 | 0.8 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 2.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 1.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.4 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 2.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 3.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 1.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 3.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |