Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
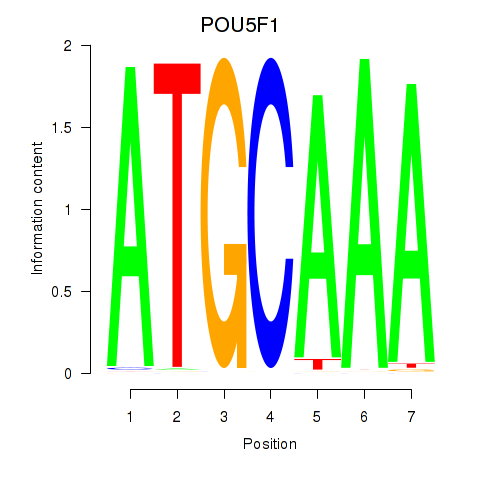
Results for POU5F1_POU2F3
Z-value: 1.85
Transcription factors associated with POU5F1_POU2F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU5F1
|
ENSG00000204531.11 | POU5F1 |
|
POU2F3
|
ENSG00000137709.5 | POU2F3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| POU2F3 | hg19_v2_chr11_+_120107344_120107351 | 0.21 | 4.3e-01 | Click! |
| POU5F1 | hg19_v2_chr6_-_31138439_31138475 | 0.06 | 8.2e-01 | Click! |
Activity profile of POU5F1_POU2F3 motif
Sorted Z-values of POU5F1_POU2F3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of POU5F1_POU2F3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_32077386 | 9.17 |
ENST00000354689.6 |
IGHV3OR16-9 |
immunoglobulin heavy variable 3/OR16-9 (non-functional) |
| chr22_+_23077065 | 7.43 |
ENST00000390310.2 |
IGLV2-18 |
immunoglobulin lambda variable 2-18 |
| chr10_-_98031310 | 6.21 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr10_-_98031265 | 6.10 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr16_+_23847267 | 5.97 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr22_+_23134974 | 5.59 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr14_-_106573756 | 5.33 |
ENST00000390601.2 |
IGHV3-11 |
immunoglobulin heavy variable 3-11 (gene/pseudogene) |
| chr16_+_23847339 | 4.91 |
ENST00000303531.7 |
PRKCB |
protein kinase C, beta |
| chr13_-_99959641 | 4.58 |
ENST00000376414.4 |
GPR183 |
G protein-coupled receptor 183 |
| chr22_+_22550113 | 4.47 |
ENST00000390285.3 |
IGLV6-57 |
immunoglobulin lambda variable 6-57 |
| chr14_-_107049312 | 3.97 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chr14_-_106552755 | 3.82 |
ENST00000390600.2 |
IGHV3-9 |
immunoglobulin heavy variable 3-9 |
| chr16_+_33605231 | 3.79 |
ENST00000570121.2 |
IGHV3OR16-12 |
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr14_-_106539557 | 3.71 |
ENST00000390599.2 |
IGHV1-8 |
immunoglobulin heavy variable 1-8 |
| chr3_-_107777208 | 3.70 |
ENST00000398258.3 |
CD47 |
CD47 molecule |
| chr22_+_23040274 | 3.70 |
ENST00000390306.2 |
IGLV2-23 |
immunoglobulin lambda variable 2-23 |
| chr14_-_106733624 | 3.43 |
ENST00000390610.2 |
IGHV1-24 |
immunoglobulin heavy variable 1-24 |
| chr4_-_71532339 | 3.37 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr8_+_56792355 | 3.19 |
ENST00000519728.1 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr14_-_106692191 | 3.15 |
ENST00000390607.2 |
IGHV3-21 |
immunoglobulin heavy variable 3-21 |
| chr3_+_98250743 | 3.10 |
ENST00000284311.3 |
GPR15 |
G protein-coupled receptor 15 |
| chr16_+_33020496 | 3.10 |
ENST00000565407.2 |
IGHV3OR16-8 |
immunoglobulin heavy variable 3/OR16-8 (non-functional) |
| chr19_-_39108568 | 3.02 |
ENST00000586296.1 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr14_-_106926724 | 2.99 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr22_+_22930626 | 2.94 |
ENST00000390302.2 |
IGLV2-33 |
immunoglobulin lambda variable 2-33 (non-functional) |
| chr19_-_39108552 | 2.93 |
ENST00000591517.1 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr8_+_56792377 | 2.90 |
ENST00000520220.2 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
| chr18_-_53089723 | 2.89 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr22_+_23165153 | 2.88 |
ENST00000390317.2 |
IGLV2-8 |
immunoglobulin lambda variable 2-8 |
| chr16_-_33647696 | 2.82 |
ENST00000558425.1 ENST00000569103.2 |
RP11-812E19.9 |
Uncharacterized protein |
| chr22_+_23101182 | 2.76 |
ENST00000390312.2 |
IGLV2-14 |
immunoglobulin lambda variable 2-14 |
| chr6_-_32784687 | 2.65 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr14_-_106845789 | 2.61 |
ENST00000390617.2 |
IGHV3-35 |
immunoglobulin heavy variable 3-35 (non-functional) |
| chr6_-_32557610 | 2.61 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr6_+_32407619 | 2.59 |
ENST00000395388.2 ENST00000374982.5 |
HLA-DRA |
major histocompatibility complex, class II, DR alpha |
| chr14_-_106963409 | 2.53 |
ENST00000390621.2 |
IGHV1-45 |
immunoglobulin heavy variable 1-45 |
| chr14_-_107078851 | 2.53 |
ENST00000390628.2 |
IGHV1-58 |
immunoglobulin heavy variable 1-58 |
| chr17_+_67410832 | 2.39 |
ENST00000590474.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr15_-_20193370 | 2.35 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr6_-_27114577 | 2.24 |
ENST00000356950.1 ENST00000396891.4 |
HIST1H2BK |
histone cluster 1, H2bk |
| chr1_-_154832316 | 2.20 |
ENST00000361147.4 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr18_-_52989525 | 2.12 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr2_+_90077680 | 2.07 |
ENST00000390270.2 |
IGKV3D-20 |
immunoglobulin kappa variable 3D-20 |
| chrX_+_154611749 | 2.07 |
ENST00000369505.3 |
F8A2 |
coagulation factor VIII-associated 2 |
| chr14_-_106994333 | 2.04 |
ENST00000390624.2 |
IGHV3-48 |
immunoglobulin heavy variable 3-48 |
| chrX_+_154114635 | 2.04 |
ENST00000369446.2 |
F8A1 |
coagulation factor VIII-associated 1 |
| chr1_+_156119798 | 2.00 |
ENST00000355014.2 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr22_+_22764088 | 2.00 |
ENST00000390299.2 |
IGLV1-40 |
immunoglobulin lambda variable 1-40 |
| chr10_-_64576105 | 1.98 |
ENST00000242480.3 ENST00000411732.1 |
EGR2 |
early growth response 2 |
| chr19_+_35820064 | 1.97 |
ENST00000341773.6 ENST00000600131.1 ENST00000270311.6 ENST00000595780.1 ENST00000597916.1 ENST00000593867.1 ENST00000600424.1 ENST00000599811.1 ENST00000536635.2 ENST00000085219.5 ENST00000544992.2 ENST00000419549.2 |
CD22 |
CD22 molecule |
| chr2_-_89399845 | 1.97 |
ENST00000479981.1 |
IGKV1-16 |
immunoglobulin kappa variable 1-16 |
| chr4_+_41258786 | 1.97 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chrX_+_80457442 | 1.95 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chr5_+_49961727 | 1.95 |
ENST00000505697.2 ENST00000503750.2 ENST00000514342.2 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr19_-_39108643 | 1.94 |
ENST00000396857.2 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr2_+_89184868 | 1.94 |
ENST00000390243.2 |
IGKV4-1 |
immunoglobulin kappa variable 4-1 |
| chrX_-_80457385 | 1.93 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr12_+_113344755 | 1.92 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr22_-_24096562 | 1.92 |
ENST00000398465.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr4_-_38806404 | 1.91 |
ENST00000308979.2 ENST00000505940.1 ENST00000515861.1 |
TLR1 |
toll-like receptor 1 |
| chr7_-_37024665 | 1.90 |
ENST00000396040.2 |
ELMO1 |
engulfment and cell motility 1 |
| chrX_+_12993202 | 1.86 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr3_-_127541194 | 1.86 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr13_-_47012325 | 1.84 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr18_-_52989217 | 1.82 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr5_-_111091948 | 1.82 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr16_+_32063311 | 1.80 |
ENST00000426099.1 |
AC142381.1 |
AC142381.1 |
| chr3_-_16524357 | 1.77 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr2_+_90139056 | 1.76 |
ENST00000492446.1 |
IGKV1D-16 |
immunoglobulin kappa variable 1D-16 |
| chr14_-_107219365 | 1.75 |
ENST00000424969.2 |
IGHV3-74 |
immunoglobulin heavy variable 3-74 |
| chr1_-_31230650 | 1.72 |
ENST00000294507.3 |
LAPTM5 |
lysosomal protein transmembrane 5 |
| chr2_+_68592305 | 1.71 |
ENST00000234313.7 |
PLEK |
pleckstrin |
| chr22_+_22712087 | 1.71 |
ENST00000390294.2 |
IGLV1-47 |
immunoglobulin lambda variable 1-47 |
| chrX_+_12993336 | 1.70 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr21_+_10862622 | 1.68 |
ENST00000302092.5 ENST00000559480.1 |
IGHV1OR21-1 |
immunoglobulin heavy variable 1/OR21-1 (non-functional) |
| chr12_+_25205568 | 1.68 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chrX_-_48776292 | 1.68 |
ENST00000376509.4 |
PIM2 |
pim-2 oncogene |
| chr12_+_14518598 | 1.66 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr14_-_106518922 | 1.65 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr11_-_73687997 | 1.65 |
ENST00000545212.1 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr2_-_175547571 | 1.65 |
ENST00000409415.3 ENST00000359761.3 ENST00000272746.5 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr12_+_113344582 | 1.64 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr12_+_25205666 | 1.64 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr13_-_99910673 | 1.57 |
ENST00000397473.2 ENST00000397470.2 |
GPR18 |
G protein-coupled receptor 18 |
| chr2_-_89521942 | 1.54 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr3_+_186648307 | 1.54 |
ENST00000457772.2 ENST00000455441.1 ENST00000427315.1 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr17_-_62009621 | 1.52 |
ENST00000349817.2 ENST00000392795.3 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
| chr7_+_116660246 | 1.49 |
ENST00000434836.1 ENST00000393443.1 ENST00000465133.1 ENST00000477742.1 ENST00000393447.4 ENST00000393444.3 |
ST7 |
suppression of tumorigenicity 7 |
| chr3_+_186648274 | 1.49 |
ENST00000169298.3 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr18_-_53257027 | 1.48 |
ENST00000568740.1 ENST00000564403.2 ENST00000537578.1 |
TCF4 |
transcription factor 4 |
| chr14_-_106471723 | 1.45 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr2_-_38303218 | 1.44 |
ENST00000407341.1 ENST00000260630.3 |
CYP1B1 |
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr16_+_33006369 | 1.43 |
ENST00000425181.3 |
IGHV3OR16-10 |
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr15_-_22448819 | 1.42 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr22_+_22735135 | 1.41 |
ENST00000390297.2 |
IGLV1-44 |
immunoglobulin lambda variable 1-44 |
| chr18_-_53255766 | 1.39 |
ENST00000566286.1 ENST00000564999.1 ENST00000566279.1 ENST00000354452.3 ENST00000356073.4 |
TCF4 |
transcription factor 4 |
| chr3_+_141106643 | 1.38 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr17_-_62009702 | 1.37 |
ENST00000006750.3 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
| chr5_+_153570285 | 1.37 |
ENST00000425427.2 ENST00000297107.6 |
GALNT10 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr12_+_25205446 | 1.36 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr22_-_24096630 | 1.35 |
ENST00000248948.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr1_+_32666188 | 1.35 |
ENST00000421922.2 |
CCDC28B |
coiled-coil domain containing 28B |
| chr11_+_313503 | 1.33 |
ENST00000528780.1 ENST00000328221.5 |
IFITM1 |
interferon induced transmembrane protein 1 |
| chr5_+_49962772 | 1.29 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr12_+_113354341 | 1.28 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_+_89998789 | 1.28 |
ENST00000453166.2 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr15_-_80263506 | 1.27 |
ENST00000335661.6 |
BCL2A1 |
BCL2-related protein A1 |
| chr10_+_11206925 | 1.26 |
ENST00000354440.2 ENST00000315874.4 ENST00000427450.1 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr2_-_89340242 | 1.25 |
ENST00000480492.1 |
IGKV1-12 |
immunoglobulin kappa variable 1-12 |
| chr6_-_24877490 | 1.24 |
ENST00000540914.1 ENST00000378023.4 |
FAM65B |
family with sequence similarity 65, member B |
| chr11_+_102188224 | 1.22 |
ENST00000263464.3 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr22_+_22385332 | 1.22 |
ENST00000390282.2 |
IGLV4-69 |
immunoglobulin lambda variable 4-69 |
| chr3_+_178253993 | 1.18 |
ENST00000420517.2 ENST00000452583.1 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
| chr7_-_37488834 | 1.18 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr7_+_64363625 | 1.18 |
ENST00000476120.1 ENST00000319636.5 ENST00000545510.1 |
ZNF273 |
zinc finger protein 273 |
| chr2_+_90153696 | 1.17 |
ENST00000417279.2 |
IGKV3D-15 |
immunoglobulin kappa variable 3D-15 (gene/pseudogene) |
| chr1_+_192544857 | 1.17 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr11_+_102188272 | 1.17 |
ENST00000532808.1 |
BIRC3 |
baculoviral IAP repeat containing 3 |
| chr14_-_107114267 | 1.16 |
ENST00000454421.2 |
IGHV3-64 |
immunoglobulin heavy variable 3-64 |
| chr14_-_106816253 | 1.13 |
ENST00000390615.2 |
IGHV3-33 |
immunoglobulin heavy variable 3-33 |
| chr12_-_102874416 | 1.11 |
ENST00000392904.1 ENST00000337514.6 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr12_-_102874378 | 1.11 |
ENST00000456098.1 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr16_-_74808710 | 1.10 |
ENST00000219368.3 ENST00000544337.1 |
FA2H |
fatty acid 2-hydroxylase |
| chr12_-_772901 | 1.09 |
ENST00000305108.4 |
NINJ2 |
ninjurin 2 |
| chr2_+_90198535 | 1.07 |
ENST00000390276.2 |
IGKV1D-12 |
immunoglobulin kappa variable 1D-12 |
| chr5_+_49962495 | 1.06 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr7_-_87856280 | 1.05 |
ENST00000490437.1 ENST00000431660.1 |
SRI |
sorcin |
| chr3_-_58200398 | 1.05 |
ENST00000318316.3 ENST00000460422.1 ENST00000483681.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr14_-_106866934 | 1.05 |
ENST00000390618.2 |
IGHV3-38 |
immunoglobulin heavy variable 3-38 (non-functional) |
| chr7_-_87856303 | 1.04 |
ENST00000394641.3 |
SRI |
sorcin |
| chr14_+_105952648 | 1.03 |
ENST00000330233.7 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr12_-_91573132 | 1.00 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr9_+_2029019 | 1.00 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_111092930 | 1.00 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr8_-_102803163 | 0.99 |
ENST00000523645.1 ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD |
neurocalcin delta |
| chr1_+_28206150 | 0.98 |
ENST00000456990.1 |
THEMIS2 |
thymocyte selection associated family member 2 |
| chr12_-_7245125 | 0.98 |
ENST00000542285.1 ENST00000540610.1 |
C1R |
complement component 1, r subcomponent |
| chr15_-_78526855 | 0.96 |
ENST00000541759.1 ENST00000558130.1 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr7_-_144435985 | 0.96 |
ENST00000549981.1 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr12_+_113344811 | 0.95 |
ENST00000551241.1 ENST00000553185.1 ENST00000550689.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr2_-_89442621 | 0.95 |
ENST00000492167.1 |
IGKV3-20 |
immunoglobulin kappa variable 3-20 |
| chr16_+_33629600 | 0.94 |
ENST00000562905.2 |
IGHV3OR16-13 |
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr4_+_68424434 | 0.93 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr20_-_7921090 | 0.92 |
ENST00000378789.3 |
HAO1 |
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr1_-_149783914 | 0.90 |
ENST00000369167.1 ENST00000427880.2 ENST00000545683.1 |
HIST2H2BF |
histone cluster 2, H2bf |
| chr12_-_7818474 | 0.90 |
ENST00000229304.4 |
APOBEC1 |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr13_-_41593425 | 0.88 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr14_-_107170409 | 0.87 |
ENST00000390633.2 |
IGHV1-69 |
immunoglobulin heavy variable 1-69 |
| chr5_-_142783694 | 0.85 |
ENST00000394466.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr21_-_35014027 | 0.84 |
ENST00000399442.1 ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chr6_-_29527702 | 0.82 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr14_-_107131560 | 0.82 |
ENST00000390632.2 |
IGHV3-66 |
immunoglobulin heavy variable 3-66 |
| chr2_+_58134756 | 0.82 |
ENST00000435505.2 ENST00000417641.2 |
VRK2 |
vaccinia related kinase 2 |
| chr14_-_107283278 | 0.81 |
ENST00000390639.2 |
IGHV7-81 |
immunoglobulin heavy variable 7-81 (non-functional) |
| chr19_+_53030906 | 0.81 |
ENST00000359798.4 ENST00000465448.1 ENST00000486474.1 ENST00000461779.1 |
ZNF808 |
zinc finger protein 808 |
| chr3_+_5229356 | 0.80 |
ENST00000256497.4 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
| chr11_-_46142948 | 0.80 |
ENST00000257821.4 |
PHF21A |
PHD finger protein 21A |
| chr19_-_17516449 | 0.80 |
ENST00000252593.6 |
BST2 |
bone marrow stromal cell antigen 2 |
| chrX_-_154688276 | 0.79 |
ENST00000369445.2 |
F8A3 |
coagulation factor VIII-associated 3 |
| chr14_-_106586656 | 0.79 |
ENST00000390602.2 |
IGHV3-13 |
immunoglobulin heavy variable 3-13 |
| chr4_+_75858318 | 0.79 |
ENST00000307428.7 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr1_-_226926864 | 0.78 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr15_-_20170354 | 0.78 |
ENST00000338912.5 |
IGHV1OR15-9 |
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr3_+_48264816 | 0.78 |
ENST00000296435.2 ENST00000576243.1 |
CAMP |
cathelicidin antimicrobial peptide |
| chr1_-_23886285 | 0.78 |
ENST00000374561.5 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
| chr1_+_79115503 | 0.77 |
ENST00000370747.4 ENST00000438486.1 ENST00000545124.1 |
IFI44 |
interferon-induced protein 44 |
| chr7_+_90338712 | 0.77 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr2_+_189839046 | 0.76 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr2_-_89597542 | 0.76 |
ENST00000465170.1 |
IGKV1-37 |
immunoglobulin kappa variable 1-37 (non-functional) |
| chrX_-_73072534 | 0.74 |
ENST00000429829.1 |
XIST |
X inactive specific transcript (non-protein coding) |
| chr18_-_53069419 | 0.74 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr12_-_102874102 | 0.73 |
ENST00000392905.2 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chr3_+_134514093 | 0.73 |
ENST00000398015.3 |
EPHB1 |
EPH receptor B1 |
| chr1_+_101702417 | 0.73 |
ENST00000305352.6 |
S1PR1 |
sphingosine-1-phosphate receptor 1 |
| chr4_+_26322409 | 0.73 |
ENST00000514807.1 ENST00000348160.4 ENST00000509158.1 ENST00000355476.3 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
| chr15_+_75080883 | 0.73 |
ENST00000567571.1 |
CSK |
c-src tyrosine kinase |
| chr14_-_106725723 | 0.72 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr19_-_41903161 | 0.71 |
ENST00000602129.1 ENST00000593771.1 ENST00000596905.1 ENST00000221233.4 |
EXOSC5 |
exosome component 5 |
| chr14_-_106453155 | 0.71 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr3_+_130650738 | 0.70 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_+_153570319 | 0.69 |
ENST00000377661.2 |
GALNT10 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr4_-_153332886 | 0.69 |
ENST00000603841.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr6_-_26032288 | 0.68 |
ENST00000244661.2 |
HIST1H3B |
histone cluster 1, H3b |
| chr7_+_64126535 | 0.68 |
ENST00000344930.3 |
ZNF107 |
zinc finger protein 107 |
| chrX_-_106960285 | 0.68 |
ENST00000503515.1 ENST00000372397.2 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr6_-_26216872 | 0.67 |
ENST00000244601.3 |
HIST1H2BG |
histone cluster 1, H2bg |
| chr12_+_54447637 | 0.67 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr6_-_143266297 | 0.67 |
ENST00000367603.2 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
| chr12_-_91576429 | 0.67 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr2_+_61108650 | 0.67 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr5_-_111092873 | 0.66 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr10_+_89622870 | 0.66 |
ENST00000371953.3 |
PTEN |
phosphatase and tensin homolog |
| chr17_-_47841485 | 0.66 |
ENST00000506156.1 ENST00000240364.2 |
FAM117A |
family with sequence similarity 117, member A |
| chr12_-_102874330 | 0.66 |
ENST00000307046.8 |
IGF1 |
insulin-like growth factor 1 (somatomedin C) |
| chrX_+_108779004 | 0.65 |
ENST00000218004.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr4_+_75858290 | 0.65 |
ENST00000513238.1 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
| chr12_-_91576561 | 0.64 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr2_+_97426631 | 0.64 |
ENST00000377075.2 |
CNNM4 |
cyclin M4 |
| chr14_+_72052983 | 0.63 |
ENST00000358550.2 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
| chr14_+_79745746 | 0.63 |
ENST00000281127.7 |
NRXN3 |
neurexin 3 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.0 | 3.0 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.9 | 48.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.8 | 5.9 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.8 | 10.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.7 | 13.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.7 | 4.6 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.6 | 1.9 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.6 | 5.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.5 | 2.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 83.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.4 | 2.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 3.8 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.4 | 6.7 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.3 | 1.0 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.3 | 0.9 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.3 | 0.9 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.3 | 0.8 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 1.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.2 | 0.7 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 0.9 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.2 | 0.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.7 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.1 | 1.9 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.8 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 2.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.5 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 1.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.5 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.3 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.1 | 2.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.3 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.1 | 3.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.1 | 0.4 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 3.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.6 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.6 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.1 | 0.5 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.9 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 1.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 2.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 1.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.1 | 0.5 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.1 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.1 | 1.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 2.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.3 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.2 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.3 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.1 | 4.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.2 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.1 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.9 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 0.2 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 1.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.3 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0001093 | TFIIB-class transcription factor binding(GO:0001093) |
| 0.0 | 0.3 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.1 | GO:0004132 | dCMP deaminase activity(GO:0004132) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.7 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 1.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 3.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.5 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.7 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.7 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.0 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.2 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 19.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.5 | 18.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 4.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 3.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 7.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 2.4 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 4.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 2.7 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.1 | 8.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.7 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.7 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.8 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.9 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 2.0 | 6.1 | GO:0070666 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 1.5 | 4.6 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 1.0 | 5.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.9 | 2.7 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.8 | 2.4 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.7 | 3.6 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.7 | 108.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.7 | 2.0 | GO:0035284 | rhombomere 3 development(GO:0021569) rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.7 | 2.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.6 | 3.6 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.5 | 1.6 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.5 | 0.9 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 1.3 | GO:1901189 | arterial endothelial cell fate commitment(GO:0060844) blood vessel lumenization(GO:0072554) positive regulation of ephrin receptor signaling pathway(GO:1901189) positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.4 | 1.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.4 | 4.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.4 | 1.4 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.3 | 1.0 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.3 | 0.3 | GO:0002312 | B cell activation involved in immune response(GO:0002312) |
| 0.3 | 0.9 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.3 | 2.1 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 3.3 | GO:0008228 | opsonization(GO:0008228) |
| 0.3 | 0.9 | GO:0046081 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.3 | 1.7 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.3 | 1.9 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.3 | 0.8 | GO:1901253 | negative regulation of dendritic cell cytokine production(GO:0002731) negative regulation of intracellular transport of viral material(GO:1901253) |
| 0.3 | 1.8 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.2 | 0.7 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.2 | 0.7 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) |
| 0.2 | 1.0 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.2 | 0.7 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 2.4 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.2 | 2.7 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.2 | 1.0 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.2 | 0.8 | GO:0036510 | trimming of terminal mannose on C branch(GO:0036510) |
| 0.2 | 4.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 1.2 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.2 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 0.7 | GO:0019249 | lactate biosynthetic process(GO:0019249) |
| 0.2 | 0.3 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 1.8 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.2 | 1.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.2 | 0.8 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 0.8 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.2 | 0.5 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.2 | 0.5 | GO:2000439 | negative regulation of eosinophil activation(GO:1902567) positive regulation of monocyte extravasation(GO:2000439) |
| 0.2 | 0.5 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 0.9 | GO:0016554 | cytidine to uridine editing(GO:0016554) DNA deamination(GO:0045006) DNA cytosine deamination(GO:0070383) |
| 0.1 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 0.7 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.1 | 0.5 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.8 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.1 | 0.6 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.6 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.1 | 0.1 | GO:0035990 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.1 | 0.5 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.1 | 0.9 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 12.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.1 | 0.3 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.1 | 0.4 | GO:1900127 | positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.1 | 17.5 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.1 | 0.1 | GO:0071902 | positive regulation of protein serine/threonine kinase activity(GO:0071902) |
| 0.1 | 0.3 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.8 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.4 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.4 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.1 | 7.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.1 | 0.7 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 1.1 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 0.4 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) negative regulation of lung blood pressure(GO:0061767) |
| 0.1 | 3.1 | GO:0072678 | T cell migration(GO:0072678) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.3 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.1 | 0.6 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 3.1 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.1 | 1.7 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.6 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.4 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.2 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.2 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.1 | 0.3 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.5 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.1 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 0.5 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.2 | GO:0042355 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.1 | 1.7 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.2 | GO:0019860 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) transformation of host cell by virus(GO:0019087) uracil metabolic process(GO:0019860) |
| 0.1 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.3 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.1 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.7 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.1 | 1.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 0.6 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.4 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.1 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.2 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.3 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.1 | 0.9 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.0 | 0.6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.6 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 1.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0032700 | negative regulation of interleukin-17 production(GO:0032700) |
| 0.0 | 1.2 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.4 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.8 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.5 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.1 | GO:0003257 | positive regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003257) positive regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901228) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0061325 | cell proliferation involved in outflow tract morphogenesis(GO:0061325) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 1.1 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.2 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.9 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.7 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.6 | GO:1903052 | positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) |
| 0.0 | 0.2 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 1.2 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.7 | GO:0048536 | spleen development(GO:0048536) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 8.6 | GO:0065004 | protein-DNA complex assembly(GO:0065004) |
| 0.0 | 0.1 | GO:0038163 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0061386 | soft palate development(GO:0060023) closure of optic fissure(GO:0061386) |
| 0.0 | 1.3 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0006226 | dUMP biosynthetic process(GO:0006226) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 4.0 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) actin filament branching(GO:0090135) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:1902474 | regulation of protein localization to synapse(GO:1902473) positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.6 | GO:0036035 | osteoclast development(GO:0036035) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.2 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 1.0 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.4 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.1 | GO:0048880 | sensory system development(GO:0048880) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.6 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) |
| 0.0 | 1.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.8 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.3 | GO:0035635 | entry of bacterium into host cell(GO:0035635) |
| 0.0 | 0.3 | GO:0043122 | regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043122) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 2.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.7 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.1 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.2 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0060067 | cervix development(GO:0060067) |
| 0.0 | 0.0 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0072660 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.2 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.4 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.6 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 1.4 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.0 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.4 | GO:0022400 | regulation of rhodopsin mediated signaling pathway(GO:0022400) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.0 | 0.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.0 | GO:0001519 | peptide amidation(GO:0001519) protein amidation(GO:0018032) peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.4 | GO:0044839 | G2/M transition of mitotic cell cycle(GO:0000086) cell cycle G2/M phase transition(GO:0044839) |
| 0.0 | 0.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.4 | 10.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.2 | 13.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 2.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 4.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 3.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 3.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 3.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 2.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 7.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.4 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 2.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 2.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.1 | 1.9 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 5.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH | Genes involved in Signaling by NOTCH |
| 0.0 | 1.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 1.0 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.1 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.1 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.9 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.8 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 2.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 1.0 | 49.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.5 | 1.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.5 | 3.6 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.4 | 2.9 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.4 | 7.8 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.3 | 4.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 2.1 | GO:0044326 | dendritic spine neck(GO:0044326) |
| 0.3 | 1.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 3.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.3 | 0.8 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.1 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.1 | 0.5 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.7 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.5 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.1 | 0.1 | GO:0097610 | cell division site(GO:0032153) cleavage furrow(GO:0032154) cell division site part(GO:0032155) cell surface furrow(GO:0097610) |
| 0.1 | 0.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.5 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.1 | 1.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.6 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.6 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.2 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.1 | 0.4 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.4 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 5.2 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.1 | 1.0 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.1 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 0.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.1 | 0.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 10.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 0.3 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 0.4 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.1 | 0.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 4.3 | GO:0070821 | tertiary granule membrane(GO:0070821) |
| 0.0 | 1.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 3.3 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.7 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 7.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.5 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 2.8 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.8 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.5 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.8 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 1.7 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 4.0 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |