Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
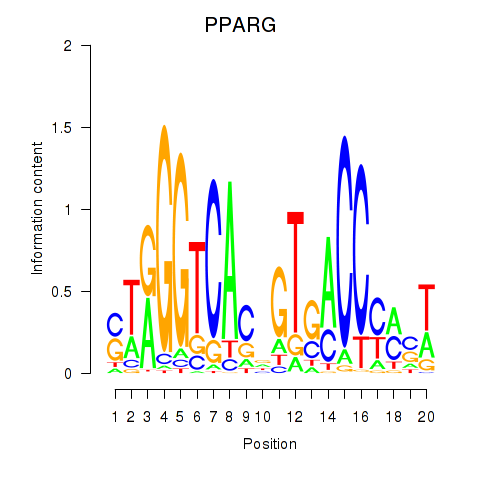
Results for PPARG
Z-value: 0.95
Transcription factors associated with PPARG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARG
|
ENSG00000132170.15 | PPARG |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PPARG | hg19_v2_chr3_+_12329397_12329433, hg19_v2_chr3_+_12392971_12392983, hg19_v2_chr3_+_12329358_12329393, hg19_v2_chr3_+_12330560_12330579 | 0.21 | 4.4e-01 | Click! |
Activity profile of PPARG motif
Sorted Z-values of PPARG motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PPARG
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_50016411 | 1.49 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr17_-_62097927 | 1.46 |
ENST00000578313.1 ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2 |
intercellular adhesion molecule 2 |
| chr17_-_62097904 | 1.38 |
ENST00000583366.1 |
ICAM2 |
intercellular adhesion molecule 2 |
| chr6_-_112575912 | 1.31 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chrX_-_51812268 | 1.23 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr6_-_112575687 | 1.10 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chrX_+_51928002 | 0.99 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr8_-_13134045 | 0.98 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr6_-_112575838 | 0.92 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr19_+_50016610 | 0.91 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_-_52441713 | 0.89 |
ENST00000182527.3 |
TRAM2 |
translocation associated membrane protein 2 |
| chr18_-_5544241 | 0.88 |
ENST00000341928.2 ENST00000540638.2 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr3_-_145968923 | 0.85 |
ENST00000493382.1 ENST00000354952.2 ENST00000383083.2 |
PLSCR4 |
phospholipid scramblase 4 |
| chr14_+_21152259 | 0.81 |
ENST00000555835.1 ENST00000336811.6 |
RNASE4 ANG |
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr11_-_9025541 | 0.80 |
ENST00000525100.1 ENST00000309166.3 ENST00000531090.1 |
NRIP3 |
nuclear receptor interacting protein 3 |
| chr3_-_145968857 | 0.79 |
ENST00000433593.2 ENST00000476202.1 ENST00000460885.1 |
PLSCR4 |
phospholipid scramblase 4 |
| chr15_+_33010175 | 0.76 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr3_+_8775466 | 0.75 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chrX_+_9433048 | 0.69 |
ENST00000217964.7 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr11_+_308143 | 0.68 |
ENST00000399817.4 |
IFITM2 |
interferon induced transmembrane protein 2 |
| chr6_-_112575758 | 0.68 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr1_+_207494853 | 0.68 |
ENST00000367064.3 ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chr19_-_13213662 | 0.68 |
ENST00000264824.4 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
| chr8_+_70404996 | 0.62 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr11_-_82612727 | 0.62 |
ENST00000531128.1 ENST00000535099.1 ENST00000527444.1 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
| chr16_+_30383613 | 0.60 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr9_-_130616915 | 0.60 |
ENST00000344849.3 |
ENG |
endoglin |
| chr21_-_46293644 | 0.58 |
ENST00000330938.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr22_+_37309662 | 0.58 |
ENST00000403662.3 ENST00000262825.5 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| chr17_-_7082861 | 0.55 |
ENST00000269299.3 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr21_-_46293586 | 0.55 |
ENST00000445724.2 ENST00000397887.3 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
| chr3_+_99357319 | 0.54 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr15_-_77376269 | 0.52 |
ENST00000558745.1 |
TSPAN3 |
tetraspanin 3 |
| chr11_+_117070037 | 0.50 |
ENST00000392951.4 ENST00000525531.1 ENST00000278968.6 |
TAGLN |
transgelin |
| chr2_+_201994042 | 0.49 |
ENST00000417748.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr4_+_15704679 | 0.47 |
ENST00000382346.3 |
BST1 |
bone marrow stromal cell antigen 1 |
| chr1_+_23037323 | 0.46 |
ENST00000544305.1 ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2 |
EPH receptor B2 |
| chr7_-_22259845 | 0.44 |
ENST00000420196.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr4_+_15704573 | 0.43 |
ENST00000265016.4 |
BST1 |
bone marrow stromal cell antigen 1 |
| chr8_-_53626974 | 0.43 |
ENST00000435644.2 ENST00000518710.1 ENST00000025008.5 ENST00000517963.1 |
RB1CC1 |
RB1-inducible coiled-coil 1 |
| chr20_+_37434329 | 0.42 |
ENST00000299824.1 ENST00000373331.2 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
| chr15_-_89089860 | 0.42 |
ENST00000558413.1 ENST00000564406.1 ENST00000268148.8 |
DET1 |
de-etiolated homolog 1 (Arabidopsis) |
| chr17_+_39975544 | 0.42 |
ENST00000544340.1 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr17_-_76899275 | 0.41 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr2_+_201994208 | 0.41 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr19_-_51587502 | 0.41 |
ENST00000156499.2 ENST00000391802.1 |
KLK14 |
kallikrein-related peptidase 14 |
| chr11_+_46740730 | 0.41 |
ENST00000311907.5 ENST00000530231.1 ENST00000442468.1 |
F2 |
coagulation factor II (thrombin) |
| chr6_+_83903061 | 0.41 |
ENST00000369724.4 ENST00000539997.1 |
RWDD2A |
RWD domain containing 2A |
| chr7_+_75932863 | 0.40 |
ENST00000429938.1 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr12_+_57849048 | 0.39 |
ENST00000266646.2 |
INHBE |
inhibin, beta E |
| chr6_+_31982539 | 0.38 |
ENST00000435363.2 ENST00000425700.2 |
C4B |
complement component 4B (Chido blood group) |
| chr19_-_48894762 | 0.38 |
ENST00000600980.1 ENST00000330720.2 |
KDELR1 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr6_+_31949801 | 0.38 |
ENST00000428956.2 ENST00000498271.1 |
C4A |
complement component 4A (Rodgers blood group) |
| chrX_+_153770421 | 0.37 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr19_-_43702231 | 0.35 |
ENST00000597374.1 ENST00000599371.1 |
PSG4 |
pregnancy specific beta-1-glycoprotein 4 |
| chr1_-_95392635 | 0.34 |
ENST00000538964.1 ENST00000394202.4 ENST00000370206.4 |
CNN3 |
calponin 3, acidic |
| chrX_+_152907913 | 0.33 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chr8_-_11725549 | 0.33 |
ENST00000505496.2 ENST00000534636.1 ENST00000524500.1 ENST00000345125.3 ENST00000453527.2 ENST00000527215.2 ENST00000532392.1 ENST00000533455.1 ENST00000534510.1 ENST00000530640.2 ENST00000531089.1 ENST00000532656.2 ENST00000531502.1 ENST00000434271.1 ENST00000353047.6 |
CTSB |
cathepsin B |
| chr11_-_5255861 | 0.32 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chr16_+_20912382 | 0.32 |
ENST00000396052.2 |
LYRM1 |
LYR motif containing 1 |
| chr17_+_45286706 | 0.31 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr2_-_69664586 | 0.31 |
ENST00000303698.3 ENST00000394305.1 ENST00000410022.2 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr19_+_11658655 | 0.30 |
ENST00000588935.1 |
CNN1 |
calponin 1, basic, smooth muscle |
| chr16_+_20912075 | 0.30 |
ENST00000219168.4 |
LYRM1 |
LYR motif containing 1 |
| chr2_+_219283815 | 0.30 |
ENST00000248444.5 ENST00000454069.1 ENST00000392114.2 |
VIL1 |
villin 1 |
| chr1_-_151762943 | 0.30 |
ENST00000368825.3 ENST00000368823.1 ENST00000458431.2 ENST00000368827.6 ENST00000368824.3 |
TDRKH |
tudor and KH domain containing |
| chr7_-_99569468 | 0.28 |
ENST00000419575.1 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr19_-_50143452 | 0.28 |
ENST00000246792.3 |
RRAS |
related RAS viral (r-ras) oncogene homolog |
| chr11_+_75526212 | 0.27 |
ENST00000356136.3 |
UVRAG |
UV radiation resistance associated |
| chr8_-_101348408 | 0.27 |
ENST00000519527.1 ENST00000522369.1 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr6_+_46097711 | 0.27 |
ENST00000321037.4 |
ENPP4 |
ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative) |
| chr15_+_84904525 | 0.27 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr10_+_114135952 | 0.27 |
ENST00000356116.1 ENST00000433418.1 ENST00000354273.4 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
| chr2_-_197664366 | 0.26 |
ENST00000409364.3 ENST00000263956.3 |
GTF3C3 |
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr11_-_5255696 | 0.25 |
ENST00000292901.3 ENST00000417377.1 |
HBD |
hemoglobin, delta |
| chr5_+_32710736 | 0.25 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr9_-_35112376 | 0.24 |
ENST00000488109.2 |
FAM214B |
family with sequence similarity 214, member B |
| chr15_-_40331342 | 0.24 |
ENST00000559081.1 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr14_+_95047725 | 0.24 |
ENST00000554760.1 ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr6_+_33378738 | 0.23 |
ENST00000374512.3 ENST00000374516.3 |
PHF1 |
PHD finger protein 1 |
| chr11_-_1593150 | 0.23 |
ENST00000397374.3 |
DUSP8 |
dual specificity phosphatase 8 |
| chr19_+_35629702 | 0.23 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr7_+_150759634 | 0.23 |
ENST00000392826.2 ENST00000461735.1 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr22_-_21579843 | 0.23 |
ENST00000405188.4 |
GGT2 |
gamma-glutamyltransferase 2 |
| chr17_+_57784826 | 0.22 |
ENST00000262291.4 |
VMP1 |
vacuole membrane protein 1 |
| chr3_-_133748913 | 0.22 |
ENST00000310926.4 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
| chr14_+_100848311 | 0.22 |
ENST00000542471.2 |
WDR25 |
WD repeat domain 25 |
| chr19_+_35630022 | 0.22 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr2_-_69664549 | 0.22 |
ENST00000450796.2 ENST00000484177.1 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr12_-_56123444 | 0.21 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr12_-_96390063 | 0.21 |
ENST00000541929.1 |
HAL |
histidine ammonia-lyase |
| chr1_+_207925391 | 0.21 |
ENST00000358170.2 ENST00000354848.1 ENST00000322918.5 ENST00000367042.1 ENST00000367041.1 ENST00000357714.1 ENST00000322875.4 ENST00000367047.1 ENST00000441839.2 ENST00000361067.1 ENST00000360212.2 ENST00000480003.1 |
CD46 |
CD46 molecule, complement regulatory protein |
| chr17_+_57784997 | 0.20 |
ENST00000537567.1 ENST00000539763.1 ENST00000587945.1 ENST00000536180.1 ENST00000589823.2 ENST00000592106.1 ENST00000591315.1 ENST00000545362.1 |
VMP1 |
vacuole membrane protein 1 |
| chr22_-_50312052 | 0.20 |
ENST00000330817.6 |
ALG12 |
ALG12, alpha-1,6-mannosyltransferase |
| chr16_+_85646891 | 0.20 |
ENST00000393243.1 |
GSE1 |
Gse1 coiled-coil protein |
| chr12_-_56122761 | 0.20 |
ENST00000552164.1 ENST00000420846.3 ENST00000257857.4 |
CD63 |
CD63 molecule |
| chr17_-_7082668 | 0.20 |
ENST00000573083.1 ENST00000574388.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr3_+_148415571 | 0.19 |
ENST00000497524.1 ENST00000349243.3 ENST00000542281.1 ENST00000418473.2 ENST00000404754.2 |
AGTR1 |
angiotensin II receptor, type 1 |
| chrX_+_54947229 | 0.19 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr12_-_96390108 | 0.19 |
ENST00000538703.1 ENST00000261208.3 |
HAL |
histidine ammonia-lyase |
| chr10_+_47894572 | 0.19 |
ENST00000355876.5 |
FAM21B |
family with sequence similarity 21, member B |
| chr19_-_46285646 | 0.19 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr2_+_30369807 | 0.18 |
ENST00000379520.3 ENST00000379519.3 ENST00000261353.4 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr11_-_67210930 | 0.18 |
ENST00000453768.2 ENST00000545016.1 ENST00000341356.5 |
CORO1B |
coronin, actin binding protein, 1B |
| chrX_+_153775821 | 0.17 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr1_-_154164534 | 0.17 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr22_+_24115000 | 0.17 |
ENST00000215743.3 |
MMP11 |
matrix metallopeptidase 11 (stromelysin 3) |
| chr15_-_40331115 | 0.17 |
ENST00000558720.1 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr12_-_80084862 | 0.17 |
ENST00000328827.4 |
PAWR |
PRKC, apoptosis, WT1, regulator |
| chr11_+_61522844 | 0.17 |
ENST00000265460.5 |
MYRF |
myelin regulatory factor |
| chr8_-_25902876 | 0.17 |
ENST00000520164.1 |
EBF2 |
early B-cell factor 2 |
| chr6_+_139094657 | 0.17 |
ENST00000332797.6 |
CCDC28A |
coiled-coil domain containing 28A |
| chr6_-_83903600 | 0.17 |
ENST00000506587.1 ENST00000507554.1 |
PGM3 |
phosphoglucomutase 3 |
| chr15_-_40331356 | 0.16 |
ENST00000560773.1 ENST00000267884.6 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
| chr1_-_161147275 | 0.16 |
ENST00000319769.5 ENST00000367998.1 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr5_+_175976324 | 0.16 |
ENST00000261944.5 |
CDHR2 |
cadherin-related family member 2 |
| chr9_-_139927462 | 0.16 |
ENST00000314412.6 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
| chrX_+_135252050 | 0.16 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr22_-_24641027 | 0.16 |
ENST00000398292.3 ENST00000263112.7 ENST00000418439.2 ENST00000424217.1 ENST00000327365.4 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr17_-_79196799 | 0.16 |
ENST00000269392.4 |
AZI1 |
5-azacytidine induced 1 |
| chr3_+_128968437 | 0.16 |
ENST00000314797.6 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
| chr7_+_73868120 | 0.15 |
ENST00000265755.3 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr17_+_48172639 | 0.15 |
ENST00000503176.1 ENST00000503614.1 |
PDK2 |
pyruvate dehydrogenase kinase, isozyme 2 |
| chr9_-_130679257 | 0.15 |
ENST00000361444.3 ENST00000335791.5 ENST00000343609.2 |
ST6GALNAC4 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4 |
| chr7_+_150498610 | 0.15 |
ENST00000461345.1 |
TMEM176A |
transmembrane protein 176A |
| chr8_+_21911054 | 0.15 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr16_+_476379 | 0.15 |
ENST00000434585.1 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
| chr11_+_844406 | 0.15 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr7_+_150498783 | 0.14 |
ENST00000475536.1 ENST00000468689.1 |
TMEM176A |
transmembrane protein 176A |
| chr11_-_6440283 | 0.14 |
ENST00000299402.6 ENST00000609360.1 ENST00000389906.2 ENST00000532020.2 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr3_+_183873098 | 0.14 |
ENST00000313143.3 |
DVL3 |
dishevelled segment polarity protein 3 |
| chr1_+_201798269 | 0.14 |
ENST00000361565.4 |
IPO9 |
importin 9 |
| chr5_+_78407602 | 0.14 |
ENST00000274353.5 ENST00000524080.1 |
BHMT |
betaine--homocysteine S-methyltransferase |
| chr17_-_74236382 | 0.14 |
ENST00000592271.1 ENST00000319945.6 ENST00000269391.6 |
RNF157 |
ring finger protein 157 |
| chr12_+_112451120 | 0.13 |
ENST00000261735.3 ENST00000455836.1 |
ERP29 |
endoplasmic reticulum protein 29 |
| chr22_+_24823517 | 0.13 |
ENST00000496258.1 ENST00000337539.7 |
ADORA2A |
adenosine A2a receptor |
| chr19_-_45826125 | 0.13 |
ENST00000221476.3 |
CKM |
creatine kinase, muscle |
| chr19_-_1592828 | 0.13 |
ENST00000592012.1 |
MBD3 |
methyl-CpG binding domain protein 3 |
| chr22_-_19974616 | 0.13 |
ENST00000344269.3 ENST00000401994.1 ENST00000406522.1 |
ARVCF |
armadillo repeat gene deleted in velocardiofacial syndrome |
| chr7_-_100425112 | 0.13 |
ENST00000358173.3 |
EPHB4 |
EPH receptor B4 |
| chr1_+_158323755 | 0.13 |
ENST00000368157.1 ENST00000368156.1 ENST00000368155.3 ENST00000368154.1 ENST00000368160.3 ENST00000368161.3 |
CD1E |
CD1e molecule |
| chr20_-_33999766 | 0.13 |
ENST00000349714.5 ENST00000438533.1 ENST00000359226.2 ENST00000374384.2 ENST00000374377.5 ENST00000407996.2 ENST00000424405.1 ENST00000542501.1 ENST00000397554.1 ENST00000540457.1 ENST00000374380.2 ENST00000374385.5 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr7_+_73868220 | 0.12 |
ENST00000455841.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr14_+_105953246 | 0.12 |
ENST00000392531.3 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr1_-_1009683 | 0.12 |
ENST00000453464.2 |
RNF223 |
ring finger protein 223 |
| chrX_-_48980098 | 0.12 |
ENST00000156109.5 |
GPKOW |
G patch domain and KOW motifs |
| chr20_+_52824367 | 0.12 |
ENST00000371419.2 |
PFDN4 |
prefoldin subunit 4 |
| chr11_+_7506713 | 0.12 |
ENST00000329293.3 ENST00000534244.1 |
OLFML1 |
olfactomedin-like 1 |
| chr19_+_51645556 | 0.12 |
ENST00000601682.1 ENST00000317643.6 ENST00000305628.7 ENST00000600577.1 |
SIGLEC7 |
sialic acid binding Ig-like lectin 7 |
| chr9_-_127263265 | 0.12 |
ENST00000373587.3 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
| chr16_+_85646763 | 0.12 |
ENST00000411612.1 ENST00000253458.7 |
GSE1 |
Gse1 coiled-coil protein |
| chr1_+_156096336 | 0.11 |
ENST00000504687.1 ENST00000473598.2 |
LMNA |
lamin A/C |
| chr17_-_56032602 | 0.11 |
ENST00000577840.1 |
CUEDC1 |
CUE domain containing 1 |
| chr17_-_2304365 | 0.11 |
ENST00000575394.1 ENST00000174618.4 |
MNT |
MAX network transcriptional repressor |
| chr22_+_19929130 | 0.11 |
ENST00000361682.6 ENST00000403184.1 ENST00000403710.1 ENST00000407537.1 |
COMT |
catechol-O-methyltransferase |
| chr16_-_67969888 | 0.11 |
ENST00000574576.2 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr18_-_55470320 | 0.11 |
ENST00000536015.1 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr11_-_65686586 | 0.11 |
ENST00000438576.2 |
C11orf68 |
chromosome 11 open reading frame 68 |
| chr8_+_29952914 | 0.11 |
ENST00000321250.8 ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
| chr11_-_117698787 | 0.11 |
ENST00000260287.2 |
FXYD2 |
FXYD domain containing ion transport regulator 2 |
| chr20_+_62496596 | 0.11 |
ENST00000369927.4 ENST00000346249.4 ENST00000348257.5 ENST00000352482.4 ENST00000351424.4 ENST00000217121.5 ENST00000358548.4 |
TPD52L2 |
tumor protein D52-like 2 |
| chr22_+_40390930 | 0.11 |
ENST00000333407.6 |
FAM83F |
family with sequence similarity 83, member F |
| chr4_-_53525406 | 0.11 |
ENST00000451218.2 ENST00000441222.3 |
USP46 |
ubiquitin specific peptidase 46 |
| chr1_+_36690011 | 0.11 |
ENST00000354618.5 ENST00000469141.2 ENST00000478853.1 |
THRAP3 |
thyroid hormone receptor associated protein 3 |
| chr14_+_105953204 | 0.10 |
ENST00000409393.2 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
| chr20_+_54933971 | 0.10 |
ENST00000371384.3 ENST00000437418.1 |
FAM210B |
family with sequence similarity 210, member B |
| chr20_+_34824355 | 0.10 |
ENST00000397286.3 ENST00000320849.4 ENST00000373932.3 |
AAR2 |
AAR2 splicing factor homolog (S. cerevisiae) |
| chr3_+_195447738 | 0.10 |
ENST00000447234.2 ENST00000320736.6 ENST00000436408.1 |
MUC20 |
mucin 20, cell surface associated |
| chr19_-_54746600 | 0.10 |
ENST00000245621.5 ENST00000270464.5 ENST00000419410.2 ENST00000391735.3 ENST00000396365.2 ENST00000440558.2 ENST00000407860.2 |
LILRA6 LILRB3 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 |
| chr9_-_130966497 | 0.10 |
ENST00000393608.1 ENST00000372948.3 |
CIZ1 |
CDKN1A interacting zinc finger protein 1 |
| chr1_-_179112189 | 0.10 |
ENST00000512653.1 ENST00000344730.3 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr3_+_52811596 | 0.10 |
ENST00000542827.1 ENST00000273283.2 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr15_+_43425672 | 0.10 |
ENST00000260403.2 |
TMEM62 |
transmembrane protein 62 |
| chr19_+_532049 | 0.10 |
ENST00000606136.1 |
CDC34 |
cell division cycle 34 |
| chr11_-_65686496 | 0.10 |
ENST00000449692.3 |
C11orf68 |
chromosome 11 open reading frame 68 |
| chr12_-_80084594 | 0.09 |
ENST00000548426.1 |
PAWR |
PRKC, apoptosis, WT1, regulator |
| chr16_+_90089008 | 0.09 |
ENST00000268699.4 |
GAS8 |
growth arrest-specific 8 |
| chr19_-_5839721 | 0.09 |
ENST00000286955.5 ENST00000318336.4 |
FUT6 |
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr6_-_31782813 | 0.09 |
ENST00000375654.4 |
HSPA1L |
heat shock 70kDa protein 1-like |
| chr6_+_31465849 | 0.09 |
ENST00000399150.3 |
MICB |
MHC class I polypeptide-related sequence B |
| chr19_-_5839703 | 0.09 |
ENST00000524754.1 |
FUT6 |
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr5_+_175223313 | 0.09 |
ENST00000359546.4 |
CPLX2 |
complexin 2 |
| chr1_+_16083154 | 0.09 |
ENST00000375771.1 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr17_-_1733114 | 0.09 |
ENST00000305513.7 |
SMYD4 |
SET and MYND domain containing 4 |
| chr11_+_14665373 | 0.09 |
ENST00000455098.2 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
| chr3_+_9958758 | 0.09 |
ENST00000383812.4 ENST00000438091.1 ENST00000295981.3 ENST00000436503.1 ENST00000403601.3 ENST00000416074.2 ENST00000455057.1 |
IL17RC |
interleukin 17 receptor C |
| chr3_+_9958870 | 0.09 |
ENST00000413608.1 |
IL17RC |
interleukin 17 receptor C |
| chr19_+_531713 | 0.09 |
ENST00000215574.4 |
CDC34 |
cell division cycle 34 |
| chr19_-_14228541 | 0.09 |
ENST00000590853.1 ENST00000308677.4 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
| chr1_+_236849754 | 0.09 |
ENST00000542672.1 ENST00000366578.4 |
ACTN2 |
actinin, alpha 2 |
| chr6_-_74161977 | 0.09 |
ENST00000370318.1 ENST00000370315.3 |
MB21D1 |
Mab-21 domain containing 1 |
| chr12_-_56583332 | 0.08 |
ENST00000347471.4 ENST00000267064.4 ENST00000394023.3 |
SMARCC2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr8_-_97247759 | 0.08 |
ENST00000518406.1 ENST00000523920.1 ENST00000287022.5 |
UQCRB |
ubiquinol-cytochrome c reductase binding protein |
| chr6_+_31465892 | 0.08 |
ENST00000252229.6 ENST00000427115.1 |
MICB |
MHC class I polypeptide-related sequence B |
| chr1_+_10459111 | 0.08 |
ENST00000541529.1 ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD |
phosphogluconate dehydrogenase |
| chr12_-_56583243 | 0.08 |
ENST00000550164.1 |
SMARCC2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr1_+_168148273 | 0.08 |
ENST00000367830.3 |
TIPRL |
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr4_-_25865159 | 0.08 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr19_-_1592652 | 0.08 |
ENST00000156825.1 ENST00000434436.3 |
MBD3 |
methyl-CpG binding domain protein 3 |
| chr20_+_44563267 | 0.08 |
ENST00000372409.3 |
PCIF1 |
PDX1 C-terminal inhibiting factor 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.3 | 0.8 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.2 | 0.6 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 0.6 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.2 | 0.7 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 0.8 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.6 | GO:0002353 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) kinin cascade(GO:0002254) plasma kallikrein-kinin cascade(GO:0002353) |
| 0.1 | 0.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.4 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.7 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 0.4 | GO:0019556 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.9 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 1.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.9 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.1 | 0.9 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 0.4 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 0.4 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.6 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.7 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.5 | GO:1904783 | positive regulation of NMDA glutamate receptor activity(GO:1904783) |
| 0.1 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.3 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.1 | 1.0 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 0.2 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.1 | 0.3 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.8 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 0.2 | GO:0090675 | intermicrovillar adhesion(GO:0090675) |
| 0.1 | 0.2 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.2 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 1.5 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.0 | 0.2 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 1.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.3 | GO:0042791 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.4 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 4.3 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.4 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.3 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:1901093 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.3 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.2 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.3 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.0 | 0.1 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.0 | GO:0055073 | cadmium ion homeostasis(GO:0055073) |
| 0.0 | 0.9 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.0 | GO:0006522 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.3 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.3 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 2.6 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.2 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.5 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.0 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.6 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.2 | GO:0036376 | sodium ion export from cell(GO:0036376) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.1 | GO:0030581 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.2 | 3.7 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.8 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 3.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.2 | GO:0097183 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.5 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 0.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 0.2 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.9 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.3 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.6 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.5 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.5 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.6 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 2.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.9 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.4 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.9 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 2.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.3 | 0.9 | GO:0050135 | NAD+ nucleosidase activity(GO:0003953) NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.2 | 0.6 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.2 | 0.6 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.8 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 0.4 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 0.6 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.8 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.4 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.4 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.1 | 0.7 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.1 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.3 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.1 | 0.6 | GO:0036122 | galactose binding(GO:0005534) BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.6 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 4.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.6 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.4 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.2 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 1.0 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 2.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.0 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.3 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.2 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |