Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
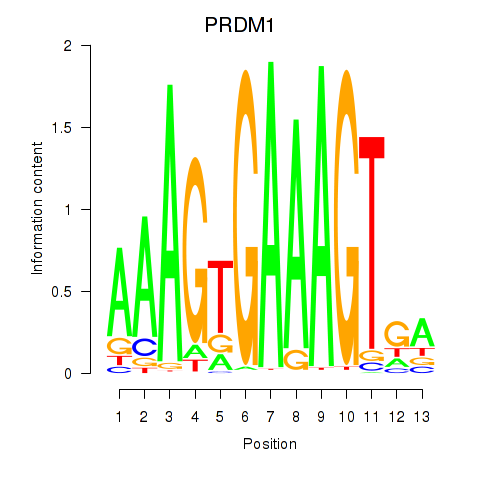
Results for PRDM1
Z-value: 1.38
Transcription factors associated with PRDM1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM1
|
ENSG00000057657.10 | PRDM1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM1 | hg19_v2_chr6_+_106534192_106534224, hg19_v2_chr6_+_106546808_106546833 | 0.01 | 9.8e-01 | Click! |
Activity profile of PRDM1 motif
Sorted Z-values of PRDM1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_112575912 | 4.27 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr6_-_112575687 | 3.31 |
ENST00000521398.1 ENST00000424408.2 ENST00000243219.3 |
LAMA4 |
laminin, alpha 4 |
| chr6_-_112575838 | 2.78 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr15_-_37393406 | 2.44 |
ENST00000338564.5 ENST00000558313.1 ENST00000340545.5 |
MEIS2 |
Meis homeobox 2 |
| chr11_+_46402583 | 2.36 |
ENST00000359803.3 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr5_+_92919043 | 2.30 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr6_-_112575758 | 2.15 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chr11_+_46402744 | 2.00 |
ENST00000533952.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr9_+_137533615 | 1.93 |
ENST00000371817.3 |
COL5A1 |
collagen, type V, alpha 1 |
| chr8_+_97506033 | 1.92 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr11_+_46402297 | 1.85 |
ENST00000405308.2 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr11_+_46402482 | 1.80 |
ENST00000441869.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr3_+_8543393 | 1.72 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr10_-_44880491 | 1.70 |
ENST00000374426.2 ENST00000395795.4 ENST00000395794.2 ENST00000374429.2 ENST00000395793.3 ENST00000343575.6 |
CXCL12 |
chemokine (C-X-C motif) ligand 12 |
| chr1_+_212782012 | 1.67 |
ENST00000341491.4 ENST00000366985.1 |
ATF3 |
activating transcription factor 3 |
| chr3_+_8543561 | 1.51 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr4_-_100242549 | 1.22 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr22_-_36556821 | 1.08 |
ENST00000531095.1 ENST00000397293.2 ENST00000349314.2 |
APOL3 |
apolipoprotein L, 3 |
| chrX_-_118827333 | 1.07 |
ENST00000360156.7 ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6 |
septin 6 |
| chr16_-_67970990 | 1.07 |
ENST00000358514.4 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr3_-_114343039 | 1.04 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr13_+_102104980 | 1.03 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr5_-_94417339 | 1.03 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr6_-_167275991 | 1.00 |
ENST00000510118.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr18_-_53070913 | 0.99 |
ENST00000568186.1 ENST00000564228.1 |
TCF4 |
transcription factor 4 |
| chr5_-_96143796 | 0.94 |
ENST00000296754.3 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr7_+_134832808 | 0.93 |
ENST00000275767.3 |
TMEM140 |
transmembrane protein 140 |
| chr6_-_29527702 | 0.92 |
ENST00000377050.4 |
UBD |
ubiquitin D |
| chr3_-_49851313 | 0.91 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr15_-_37392086 | 0.88 |
ENST00000561208.1 |
MEIS2 |
Meis homeobox 2 |
| chr5_-_93447333 | 0.88 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr12_+_51318513 | 0.87 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr3_-_46000064 | 0.86 |
ENST00000433878.1 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr6_-_167276033 | 0.82 |
ENST00000503859.1 ENST00000506565.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chrX_-_119694538 | 0.82 |
ENST00000371322.5 |
CUL4B |
cullin 4B |
| chr4_+_142558078 | 0.81 |
ENST00000529613.1 |
IL15 |
interleukin 15 |
| chr8_-_29120580 | 0.81 |
ENST00000524189.1 |
KIF13B |
kinesin family member 13B |
| chr4_+_142557771 | 0.80 |
ENST00000514653.1 |
IL15 |
interleukin 15 |
| chr2_-_145275228 | 0.79 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr17_-_26694979 | 0.78 |
ENST00000438614.1 |
VTN |
vitronectin |
| chr1_+_207262881 | 0.77 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr17_-_26695013 | 0.77 |
ENST00000555059.2 |
CTB-96E2.2 |
Homeobox protein SEBOX |
| chr7_-_86849025 | 0.76 |
ENST00000257637.3 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr14_+_24605389 | 0.76 |
ENST00000382708.3 ENST00000561435.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr4_+_142557717 | 0.75 |
ENST00000320650.4 ENST00000296545.7 |
IL15 |
interleukin 15 |
| chr3_+_69915385 | 0.75 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr3_+_8543533 | 0.72 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr21_-_40033618 | 0.65 |
ENST00000417133.2 ENST00000398910.1 ENST00000442448.1 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr14_+_24605361 | 0.64 |
ENST00000206451.6 ENST00000559123.1 |
PSME1 |
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr17_+_41158742 | 0.63 |
ENST00000415816.2 ENST00000438323.2 |
IFI35 |
interferon-induced protein 35 |
| chr11_+_128634589 | 0.62 |
ENST00000281428.8 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_-_122526499 | 0.60 |
ENST00000412584.2 |
CADPS2 |
Ca++-dependent secretion activator 2 |
| chr20_+_30946106 | 0.54 |
ENST00000375687.4 ENST00000542461.1 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
| chr3_+_159706537 | 0.54 |
ENST00000305579.2 ENST00000480787.1 ENST00000466512.1 |
IL12A |
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr11_+_128563948 | 0.53 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr9_-_117568365 | 0.49 |
ENST00000374045.4 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
| chr12_-_106641728 | 0.49 |
ENST00000378026.4 |
CKAP4 |
cytoskeleton-associated protein 4 |
| chr6_-_32811771 | 0.47 |
ENST00000395339.3 ENST00000374882.3 |
PSMB8 |
proteasome (prosome, macropain) subunit, beta type, 8 |
| chr6_+_32811885 | 0.47 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_+_231191875 | 0.44 |
ENST00000444636.1 ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L |
SP140 nuclear body protein-like |
| chr12_-_71003568 | 0.43 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr1_-_153919128 | 0.40 |
ENST00000361217.4 |
DENND4B |
DENN/MADD domain containing 4B |
| chr12_+_94542459 | 0.38 |
ENST00000258526.4 |
PLXNC1 |
plexin C1 |
| chr6_-_137113604 | 0.38 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr1_+_28099683 | 0.37 |
ENST00000373943.4 |
STX12 |
syntaxin 12 |
| chr4_-_100140331 | 0.36 |
ENST00000407820.2 ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6 |
alcohol dehydrogenase 6 (class V) |
| chrX_-_154563889 | 0.36 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
| chr12_-_54653313 | 0.36 |
ENST00000550411.1 ENST00000439541.2 |
CBX5 |
chromobox homolog 5 |
| chr11_+_128563652 | 0.35 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr2_-_64246206 | 0.34 |
ENST00000409558.4 ENST00000272322.4 |
VPS54 |
vacuolar protein sorting 54 homolog (S. cerevisiae) |
| chr20_+_388791 | 0.34 |
ENST00000441733.1 ENST00000353660.3 |
RBCK1 |
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr5_-_96143602 | 0.34 |
ENST00000443439.2 ENST00000503921.1 ENST00000508227.1 ENST00000507154.1 |
ERAP1 |
endoplasmic reticulum aminopeptidase 1 |
| chr6_+_32821924 | 0.33 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_+_146403912 | 0.31 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chr11_+_64323428 | 0.30 |
ENST00000377581.3 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr19_-_6670128 | 0.29 |
ENST00000245912.3 |
TNFSF14 |
tumor necrosis factor (ligand) superfamily, member 14 |
| chr6_+_31540056 | 0.29 |
ENST00000418386.2 |
LTA |
lymphotoxin alpha |
| chr17_-_79818354 | 0.28 |
ENST00000576541.1 ENST00000576380.1 ENST00000571617.1 ENST00000576052.1 ENST00000576390.1 ENST00000573778.2 ENST00000439918.2 ENST00000574914.1 ENST00000331483.4 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr2_-_175260368 | 0.27 |
ENST00000342016.3 ENST00000362053.5 |
CIR1 |
corepressor interacting with RBPJ, 1 |
| chr1_+_110453203 | 0.27 |
ENST00000357302.4 ENST00000344188.5 ENST00000329608.6 |
CSF1 |
colony stimulating factor 1 (macrophage) |
| chr14_-_24615805 | 0.27 |
ENST00000560410.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chr17_+_46908350 | 0.26 |
ENST00000258947.3 ENST00000509507.1 ENST00000448105.2 ENST00000570513.1 ENST00000509415.1 ENST00000513119.1 ENST00000416445.2 ENST00000508679.1 ENST00000505071.1 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
| chr12_+_94656297 | 0.25 |
ENST00000545312.1 |
PLXNC1 |
plexin C1 |
| chr12_+_6561190 | 0.25 |
ENST00000544021.1 ENST00000266556.7 |
TAPBPL |
TAP binding protein-like |
| chr22_+_41697520 | 0.25 |
ENST00000352645.4 |
ZC3H7B |
zinc finger CCCH-type containing 7B |
| chr8_-_56986768 | 0.23 |
ENST00000523936.1 |
RPS20 |
ribosomal protein S20 |
| chr11_+_64323098 | 0.21 |
ENST00000301891.4 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr2_+_231280908 | 0.21 |
ENST00000427101.2 ENST00000432979.1 |
SP100 |
SP100 nuclear antigen |
| chr19_-_43032532 | 0.21 |
ENST00000403461.1 ENST00000352591.5 ENST00000358394.3 ENST00000403444.3 ENST00000308072.4 ENST00000599389.1 ENST00000351134.3 ENST00000161559.6 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
| chr3_-_168864427 | 0.21 |
ENST00000468789.1 |
MECOM |
MDS1 and EVI1 complex locus |
| chr10_-_82049424 | 0.19 |
ENST00000372213.3 |
MAT1A |
methionine adenosyltransferase I, alpha |
| chr14_-_24615523 | 0.19 |
ENST00000559056.1 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
| chrX_+_10124977 | 0.19 |
ENST00000380833.4 |
CLCN4 |
chloride channel, voltage-sensitive 4 |
| chr1_-_244006528 | 0.19 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr2_+_145780767 | 0.19 |
ENST00000599358.1 ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr19_+_17186577 | 0.18 |
ENST00000595618.1 ENST00000594824.1 |
MYO9B |
myosin IXB |
| chr16_+_30675654 | 0.18 |
ENST00000287468.5 ENST00000395073.2 |
FBRS |
fibrosin |
| chr3_-_4793274 | 0.17 |
ENST00000414938.1 |
EGOT |
eosinophil granule ontogeny transcript (non-protein coding) |
| chr1_-_111746966 | 0.16 |
ENST00000369752.5 |
DENND2D |
DENN/MADD domain containing 2D |
| chr6_+_57182400 | 0.16 |
ENST00000607273.1 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
| chr10_-_112678692 | 0.15 |
ENST00000605742.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr21_-_35899113 | 0.15 |
ENST00000492600.1 ENST00000481448.1 ENST00000381132.2 |
RCAN1 |
regulator of calcineurin 1 |
| chr22_-_36635684 | 0.15 |
ENST00000358502.5 |
APOL2 |
apolipoprotein L, 2 |
| chr19_+_49977818 | 0.15 |
ENST00000594009.1 ENST00000595510.1 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
| chr22_-_31688431 | 0.14 |
ENST00000402249.3 ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
| chr11_+_64323156 | 0.14 |
ENST00000377585.3 |
SLC22A11 |
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr3_-_149375783 | 0.13 |
ENST00000467467.1 ENST00000460517.1 ENST00000360632.3 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr13_+_31309645 | 0.12 |
ENST00000380490.3 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
| chr17_+_3379284 | 0.12 |
ENST00000263080.2 |
ASPA |
aspartoacylase |
| chr6_-_28220002 | 0.11 |
ENST00000377294.2 |
ZKSCAN4 |
zinc finger with KRAB and SCAN domains 4 |
| chr4_-_74088800 | 0.11 |
ENST00000509867.2 |
ANKRD17 |
ankyrin repeat domain 17 |
| chr1_-_205326022 | 0.10 |
ENST00000367155.3 |
KLHDC8A |
kelch domain containing 8A |
| chr16_-_3930724 | 0.09 |
ENST00000262367.5 |
CREBBP |
CREB binding protein |
| chr2_-_209010874 | 0.09 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chr11_-_72432950 | 0.09 |
ENST00000426523.1 ENST00000429686.1 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr12_+_25205446 | 0.08 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr11_+_28129795 | 0.08 |
ENST00000406787.3 ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15 |
methyltransferase like 15 |
| chr2_+_145780739 | 0.07 |
ENST00000597173.1 ENST00000602108.1 ENST00000420472.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr15_+_75491213 | 0.07 |
ENST00000360639.2 |
C15orf39 |
chromosome 15 open reading frame 39 |
| chr1_-_8483723 | 0.07 |
ENST00000476556.1 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
| chr6_+_26156551 | 0.06 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr14_-_21490417 | 0.06 |
ENST00000556366.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_23479331 | 0.05 |
ENST00000397377.1 ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93 |
chromosome 14 open reading frame 93 |
| chr19_+_13106383 | 0.05 |
ENST00000397661.2 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr6_-_121655552 | 0.05 |
ENST00000275159.6 |
TBC1D32 |
TBC1 domain family, member 32 |
| chr6_+_167525277 | 0.04 |
ENST00000400926.2 |
CCR6 |
chemokine (C-C motif) receptor 6 |
| chr22_+_39417118 | 0.04 |
ENST00000216099.8 |
APOBEC3D |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
| chr10_+_18429671 | 0.04 |
ENST00000282343.8 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr1_-_205325850 | 0.02 |
ENST00000537168.1 |
KLHDC8A |
kelch domain containing 8A |
| chr2_+_231280954 | 0.02 |
ENST00000409824.1 ENST00000409341.1 ENST00000409112.1 ENST00000340126.4 ENST00000341950.4 |
SP100 |
SP100 nuclear antigen |
| chr11_-_47736896 | 0.02 |
ENST00000525123.1 ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2 |
ATP/GTP binding protein-like 2 |
| chr8_+_134203273 | 0.02 |
ENST00000250160.6 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
| chr6_+_146348810 | 0.01 |
ENST00000492807.2 |
GRM1 |
glutamate receptor, metabotropic 1 |
| chr1_+_17906970 | 0.01 |
ENST00000375415.1 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr6_+_146348782 | 0.01 |
ENST00000361719.2 ENST00000392299.2 |
GRM1 |
glutamate receptor, metabotropic 1 |
| chr14_-_21490590 | 0.00 |
ENST00000557633.1 |
NDRG2 |
NDRG family member 2 |
| chr16_-_88851618 | 0.00 |
ENST00000301015.9 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.0 | GO:0030421 | defecation(GO:0030421) |
| 0.8 | 2.4 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.6 | 1.7 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.5 | 1.9 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 1.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.9 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 0.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.2 | 1.1 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.2 | 0.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.2 | 0.5 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.2 | 1.6 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 3.9 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.7 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.2 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.8 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) |
| 0.1 | 0.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.3 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 0.8 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.1 | 12.3 | GO:0045995 | regulation of embryonic development(GO:0045995) |
| 0.1 | 0.2 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.1 | 0.7 | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) |
| 0.1 | 0.7 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 3.3 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 4.2 | GO:0006521 | regulation of cellular amino acid metabolic process(GO:0006521) |
| 0.0 | 0.3 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 1.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.5 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.4 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.6 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.8 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 2.3 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 9.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 1.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.8 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.6 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 1.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 1.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 2.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.0 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 0.6 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 0.5 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.9 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.3 | 2.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 1.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 1.6 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 1.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 12.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 1.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 2.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 0.5 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.7 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.9 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 8.0 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 9.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 1.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.6 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.1 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.6 | 1.7 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.3 | 1.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.3 | 12.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 2.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.8 | GO:0071062 | rough endoplasmic reticulum lumen(GO:0048237) alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.1 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.3 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.7 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.5 | GO:0031906 | late endosome lumen(GO:0031906) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.4 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 1.9 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:1904724 | lipid particle(GO:0005811) tertiary granule lumen(GO:1904724) |