Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
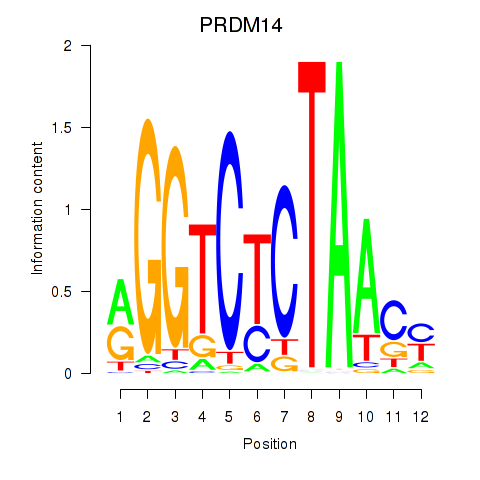
Results for PRDM14
Z-value: 0.40
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PRDM14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRDM14 | hg19_v2_chr8_-_70983506_70983562 | -0.11 | 6.7e-01 | Click! |
Activity profile of PRDM14 motif
Sorted Z-values of PRDM14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PRDM14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_23241661 | 0.96 |
ENST00000390322.2 |
IGLJ2 |
immunoglobulin lambda joining 2 |
| chr18_+_57567180 | 0.86 |
ENST00000316660.6 ENST00000269518.9 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr19_-_49314169 | 0.85 |
ENST00000597011.1 ENST00000601681.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chr4_-_103266626 | 0.83 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr19_-_49314269 | 0.80 |
ENST00000545387.2 ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2 |
branched chain amino-acid transaminase 2, mitochondrial |
| chrX_+_15525426 | 0.59 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr11_-_62474803 | 0.59 |
ENST00000533982.1 ENST00000360796.5 |
BSCL2 |
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr3_+_16926441 | 0.51 |
ENST00000418129.2 ENST00000396755.2 |
PLCL2 |
phospholipase C-like 2 |
| chr1_+_36621529 | 0.42 |
ENST00000316156.4 |
MAP7D1 |
MAP7 domain containing 1 |
| chr2_-_145278475 | 0.38 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr9_+_132597722 | 0.36 |
ENST00000372429.3 ENST00000315480.4 ENST00000358355.1 |
USP20 |
ubiquitin specific peptidase 20 |
| chr3_+_37035289 | 0.32 |
ENST00000455445.2 ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1 |
mutL homolog 1 |
| chr12_+_121647868 | 0.30 |
ENST00000359949.7 ENST00000541532.1 ENST00000543171.1 ENST00000538701.1 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr19_+_39109735 | 0.30 |
ENST00000593149.1 ENST00000248342.4 ENST00000538434.1 ENST00000588934.1 ENST00000545173.2 ENST00000589307.1 ENST00000586513.1 ENST00000591409.1 ENST00000592558.1 |
EIF3K |
eukaryotic translation initiation factor 3, subunit K |
| chr1_-_27216729 | 0.30 |
ENST00000431781.2 ENST00000374135.4 |
GPN2 |
GPN-loop GTPase 2 |
| chr3_+_37035263 | 0.30 |
ENST00000458205.2 ENST00000539477.1 |
MLH1 |
mutL homolog 1 |
| chr12_+_121647962 | 0.29 |
ENST00000542067.1 |
P2RX4 |
purinergic receptor P2X, ligand-gated ion channel, 4 |
| chr19_+_49838653 | 0.29 |
ENST00000598095.1 ENST00000426897.2 ENST00000323906.4 ENST00000535669.2 ENST00000597602.1 ENST00000595660.1 |
CD37 |
CD37 molecule |
| chr11_+_85956182 | 0.26 |
ENST00000327320.4 ENST00000351625.6 ENST00000534595.1 |
EED |
embryonic ectoderm development |
| chrX_+_41193407 | 0.25 |
ENST00000457138.2 ENST00000441189.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr12_-_49351228 | 0.25 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr14_-_21492251 | 0.23 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr12_-_49351148 | 0.22 |
ENST00000398092.4 ENST00000539611.1 |
RP11-302B13.5 ARF3 |
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr14_-_21492113 | 0.21 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr2_-_192711968 | 0.20 |
ENST00000304141.4 |
SDPR |
serum deprivation response |
| chr8_-_102216925 | 0.20 |
ENST00000517844.1 |
ZNF706 |
zinc finger protein 706 |
| chr12_-_49351303 | 0.18 |
ENST00000256682.4 |
ARF3 |
ADP-ribosylation factor 3 |
| chr19_+_3572758 | 0.17 |
ENST00000416526.1 |
HMG20B |
high mobility group 20B |
| chr1_+_65730385 | 0.15 |
ENST00000263441.7 ENST00000395325.3 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr7_-_124405681 | 0.15 |
ENST00000303921.2 |
GPR37 |
G protein-coupled receptor 37 (endothelin receptor type B-like) |
| chr2_+_113033164 | 0.14 |
ENST00000409871.1 ENST00000343936.4 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
| chr7_-_140714430 | 0.13 |
ENST00000393008.3 |
MRPS33 |
mitochondrial ribosomal protein S33 |
| chr4_+_81187753 | 0.12 |
ENST00000312465.7 ENST00000456523.3 |
FGF5 |
fibroblast growth factor 5 |
| chr5_+_32711419 | 0.12 |
ENST00000265074.8 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr7_-_140714739 | 0.11 |
ENST00000467334.1 ENST00000324787.5 |
MRPS33 |
mitochondrial ribosomal protein S33 |
| chr6_-_46048116 | 0.11 |
ENST00000185206.6 |
CLIC5 |
chloride intracellular channel 5 |
| chr2_-_69098566 | 0.10 |
ENST00000295379.1 |
BMP10 |
bone morphogenetic protein 10 |
| chr3_+_10289707 | 0.10 |
ENST00000287652.4 |
TATDN2 |
TatD DNase domain containing 2 |
| chr19_-_6279932 | 0.10 |
ENST00000252674.7 |
MLLT1 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr7_+_39989611 | 0.09 |
ENST00000181839.4 |
CDK13 |
cyclin-dependent kinase 13 |
| chr1_+_50571949 | 0.09 |
ENST00000357083.4 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr8_+_110346546 | 0.09 |
ENST00000521662.1 ENST00000521688.1 ENST00000520147.1 |
ENY2 |
enhancer of yellow 2 homolog (Drosophila) |
| chr17_-_46691990 | 0.08 |
ENST00000576562.1 |
HOXB8 |
homeobox B8 |
| chr7_-_139763521 | 0.07 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr11_+_62475130 | 0.07 |
ENST00000294117.5 |
GNG3 |
guanine nucleotide binding protein (G protein), gamma 3 |
| chr2_-_228244013 | 0.06 |
ENST00000304568.3 |
TM4SF20 |
transmembrane 4 L six family member 20 |
| chr6_-_109777128 | 0.06 |
ENST00000358807.3 ENST00000358577.3 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr5_+_32710736 | 0.06 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr17_-_46692287 | 0.06 |
ENST00000239144.4 |
HOXB8 |
homeobox B8 |
| chr1_-_116383322 | 0.03 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr1_-_42384343 | 0.03 |
ENST00000372584.1 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
| chr1_+_153963227 | 0.03 |
ENST00000368567.4 ENST00000392558.4 |
RPS27 |
ribosomal protein S27 |
| chr15_+_45694523 | 0.02 |
ENST00000305560.6 |
SPATA5L1 |
spermatogenesis associated 5-like 1 |
| chr11_-_65624415 | 0.02 |
ENST00000524553.1 ENST00000527344.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr20_-_48770174 | 0.01 |
ENST00000341698.2 |
TMEM189-UBE2V1 |
TMEM189-UBE2V1 readthrough |
| chr12_+_3600356 | 0.01 |
ENST00000382622.3 |
PRMT8 |
protein arginine methyltransferase 8 |
| chr12_+_56325812 | 0.01 |
ENST00000394147.1 ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr2_-_37193606 | 0.00 |
ENST00000379213.2 ENST00000263918.4 |
STRN |
striatin, calmodulin binding protein |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.1 | GO:0002945 | cyclin K-CDK13 complex(GO:0002945) |
| 0.0 | 0.6 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.6 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.6 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.1 | 0.6 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.2 | 0.6 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 0.5 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.1 | 0.3 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.9 | GO:0010917 | negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.1 | 0.8 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.6 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.3 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.4 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.1 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |