Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for PROX1
Z-value: 0.86
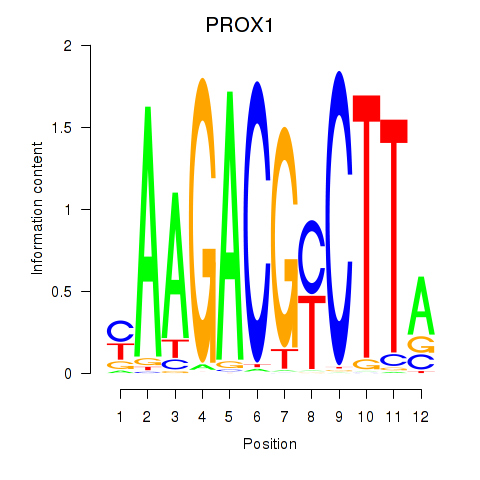
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | PROX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PROX1 | hg19_v2_chr1_+_214161272_214161322, hg19_v2_chr1_+_214161854_214161892 | 0.34 | 1.9e-01 | Click! |
Activity profile of PROX1 motif
Sorted Z-values of PROX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PROX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_22918021 | 2.24 |
ENST00000288666.5 |
RPS4Y2 |
ribosomal protein S4, Y-linked 2 |
| chr1_-_173176452 | 1.39 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr10_-_92681033 | 1.31 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr4_+_4861385 | 0.86 |
ENST00000382723.4 |
MSX1 |
msh homeobox 1 |
| chr13_-_36788718 | 0.79 |
ENST00000317764.6 ENST00000379881.3 |
SOHLH2 |
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr19_+_35773242 | 0.65 |
ENST00000222304.3 |
HAMP |
hepcidin antimicrobial peptide |
| chr6_-_7911042 | 0.61 |
ENST00000379757.4 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
| chr18_+_2571510 | 0.58 |
ENST00000261597.4 ENST00000575515.1 |
NDC80 |
NDC80 kinetochore complex component |
| chr15_-_58357932 | 0.57 |
ENST00000347587.3 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr1_+_70876891 | 0.56 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr2_+_108443388 | 0.54 |
ENST00000354986.4 ENST00000408999.3 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
| chr18_+_33767473 | 0.54 |
ENST00000261326.5 |
MOCOS |
molybdenum cofactor sulfurase |
| chr7_+_150759634 | 0.54 |
ENST00000392826.2 ENST00000461735.1 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr10_+_30722866 | 0.54 |
ENST00000263056.1 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
| chr11_-_73882029 | 0.53 |
ENST00000539061.1 |
C2CD3 |
C2 calcium-dependent domain containing 3 |
| chr11_+_46740730 | 0.52 |
ENST00000311907.5 ENST00000530231.1 ENST00000442468.1 |
F2 |
coagulation factor II (thrombin) |
| chrX_-_152486108 | 0.51 |
ENST00000356661.5 |
MAGEA1 |
melanoma antigen family A, 1 (directs expression of antigen MZ2-E) |
| chr1_-_225616515 | 0.50 |
ENST00000338179.2 ENST00000425080.1 |
LBR |
lamin B receptor |
| chr19_-_9546227 | 0.49 |
ENST00000361451.2 ENST00000361151.1 |
ZNF266 |
zinc finger protein 266 |
| chr1_-_151431909 | 0.49 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr14_+_24563262 | 0.48 |
ENST00000559250.1 ENST00000216780.4 ENST00000560736.1 ENST00000396973.4 ENST00000559837.1 |
PCK2 |
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr1_+_70876926 | 0.47 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr6_+_34725263 | 0.45 |
ENST00000374018.1 ENST00000374017.3 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr12_-_122296755 | 0.44 |
ENST00000289004.4 |
HPD |
4-hydroxyphenylpyruvate dioxygenase |
| chr11_+_1874200 | 0.43 |
ENST00000311604.3 |
LSP1 |
lymphocyte-specific protein 1 |
| chr19_-_46234119 | 0.42 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chr1_+_154966058 | 0.42 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr19_-_9546177 | 0.42 |
ENST00000592292.1 ENST00000588221.1 |
ZNF266 |
zinc finger protein 266 |
| chr22_-_39640756 | 0.42 |
ENST00000331163.6 |
PDGFB |
platelet-derived growth factor beta polypeptide |
| chrX_+_151883090 | 0.40 |
ENST00000370293.2 ENST00000423993.1 ENST00000447530.1 ENST00000458057.1 ENST00000331220.2 ENST00000422085.1 ENST00000453150.1 ENST00000409560.1 |
MAGEA2B |
melanoma antigen family A, 2B |
| chr2_-_107084826 | 0.40 |
ENST00000304514.7 ENST00000409886.3 |
RGPD3 |
RANBP2-like and GRIP domain containing 3 |
| chr5_+_156693159 | 0.40 |
ENST00000347377.6 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr11_+_61522844 | 0.38 |
ENST00000265460.5 |
MYRF |
myelin regulatory factor |
| chr9_+_131218698 | 0.38 |
ENST00000434106.3 ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr21_-_40033618 | 0.37 |
ENST00000417133.2 ENST00000398910.1 ENST00000442448.1 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
| chr18_+_60190226 | 0.36 |
ENST00000269499.5 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
| chr2_+_163175394 | 0.35 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr15_-_91537723 | 0.34 |
ENST00000394249.3 ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1 |
protein regulator of cytokinesis 1 |
| chr16_+_31885079 | 0.33 |
ENST00000300870.10 ENST00000394846.3 |
ZNF267 |
zinc finger protein 267 |
| chrX_-_151922340 | 0.33 |
ENST00000370284.1 ENST00000543232.1 ENST00000393876.1 ENST00000393872.3 |
MAGEA2 |
melanoma antigen family A, 2 |
| chr16_+_58059470 | 0.33 |
ENST00000219271.3 |
MMP15 |
matrix metallopeptidase 15 (membrane-inserted) |
| chr12_+_49740700 | 0.33 |
ENST00000549441.2 ENST00000395069.3 |
DNAJC22 |
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr17_+_26646121 | 0.33 |
ENST00000226230.6 |
TMEM97 |
transmembrane protein 97 |
| chr1_+_222817629 | 0.32 |
ENST00000340535.7 |
MIA3 |
melanoma inhibitory activity family, member 3 |
| chr14_-_23426270 | 0.32 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr5_-_180688105 | 0.32 |
ENST00000327767.4 |
TRIM52 |
tripartite motif containing 52 |
| chr18_-_21166841 | 0.32 |
ENST00000269228.5 |
NPC1 |
Niemann-Pick disease, type C1 |
| chr2_+_97001491 | 0.32 |
ENST00000240423.4 ENST00000427946.1 ENST00000435975.1 ENST00000456906.1 ENST00000455200.1 |
NCAPH |
non-SMC condensin I complex, subunit H |
| chr6_-_25042231 | 0.32 |
ENST00000510784.2 |
FAM65B |
family with sequence similarity 65, member B |
| chr13_+_37581115 | 0.31 |
ENST00000481013.1 |
EXOSC8 |
exosome component 8 |
| chr10_+_112327425 | 0.30 |
ENST00000361804.4 |
SMC3 |
structural maintenance of chromosomes 3 |
| chr14_-_23426322 | 0.30 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr17_+_55183261 | 0.30 |
ENST00000576295.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr15_-_75249793 | 0.30 |
ENST00000322177.5 |
RPP25 |
ribonuclease P/MRP 25kDa subunit |
| chr7_+_66093851 | 0.30 |
ENST00000275532.3 |
KCTD7 |
potassium channel tetramerization domain containing 7 |
| chr10_+_14920843 | 0.29 |
ENST00000433779.1 ENST00000378325.3 ENST00000354919.6 ENST00000313519.5 ENST00000420416.1 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
| chr5_+_154320623 | 0.26 |
ENST00000523037.1 ENST00000265229.8 ENST00000439747.3 ENST00000522038.1 |
MRPL22 |
mitochondrial ribosomal protein L22 |
| chr14_-_23426337 | 0.26 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr19_+_4007644 | 0.26 |
ENST00000262971.2 |
PIAS4 |
protein inhibitor of activated STAT, 4 |
| chr9_+_131267052 | 0.26 |
ENST00000539582.1 |
GLE1 |
GLE1 RNA export mediator |
| chr17_-_36904437 | 0.26 |
ENST00000585100.1 ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr9_+_93564191 | 0.25 |
ENST00000375747.1 |
SYK |
spleen tyrosine kinase |
| chr22_-_29075853 | 0.25 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chr15_+_85523671 | 0.25 |
ENST00000310298.4 ENST00000557957.1 |
PDE8A |
phosphodiesterase 8A |
| chr21_+_35736302 | 0.25 |
ENST00000290310.3 |
KCNE2 |
potassium voltage-gated channel, Isk-related family, member 2 |
| chr4_-_169931393 | 0.24 |
ENST00000504480.1 ENST00000306193.3 |
CBR4 |
carbonyl reductase 4 |
| chr1_-_159046617 | 0.23 |
ENST00000368130.4 |
AIM2 |
absent in melanoma 2 |
| chr4_+_2819883 | 0.23 |
ENST00000511747.1 ENST00000503393.2 |
SH3BP2 |
SH3-domain binding protein 2 |
| chr6_+_37321823 | 0.23 |
ENST00000487950.1 ENST00000469731.1 |
RNF8 |
ring finger protein 8, E3 ubiquitin protein ligase |
| chr19_-_15529790 | 0.23 |
ENST00000596195.1 ENST00000595067.1 ENST00000595465.2 ENST00000397410.5 ENST00000600247.1 |
AKAP8L |
A kinase (PRKA) anchor protein 8-like |
| chr8_+_110552831 | 0.23 |
ENST00000530629.1 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr7_-_102257139 | 0.22 |
ENST00000521076.1 ENST00000462172.1 ENST00000522801.1 ENST00000449970.2 ENST00000262940.7 |
RASA4 |
RAS p21 protein activator 4 |
| chr3_-_33138624 | 0.22 |
ENST00000445488.2 ENST00000307377.8 ENST00000440656.1 ENST00000436768.1 ENST00000307363.5 |
GLB1 |
galactosidase, beta 1 |
| chr10_+_16478942 | 0.22 |
ENST00000535784.2 ENST00000423462.2 ENST00000378000.1 |
PTER |
phosphotriesterase related |
| chr11_+_73882311 | 0.22 |
ENST00000398427.4 ENST00000544401.1 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr16_-_23607598 | 0.22 |
ENST00000562133.1 ENST00000570319.1 ENST00000007516.3 |
NDUFAB1 |
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa |
| chr18_-_2571437 | 0.21 |
ENST00000574676.1 ENST00000574538.1 ENST00000319888.6 |
METTL4 |
methyltransferase like 4 |
| chr11_+_73882144 | 0.21 |
ENST00000328257.8 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr1_-_15850676 | 0.21 |
ENST00000440484.1 ENST00000333868.5 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr1_-_15850839 | 0.21 |
ENST00000348549.5 ENST00000546424.1 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr3_+_152552685 | 0.21 |
ENST00000305097.3 |
P2RY1 |
purinergic receptor P2Y, G-protein coupled, 1 |
| chr1_-_28241024 | 0.21 |
ENST00000313433.7 ENST00000444045.1 |
RPA2 |
replication protein A2, 32kDa |
| chr16_+_29817399 | 0.21 |
ENST00000545521.1 |
MAZ |
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr3_+_190105909 | 0.21 |
ENST00000456423.1 |
CLDN16 |
claudin 16 |
| chr17_-_40273348 | 0.21 |
ENST00000225916.5 |
KAT2A |
K(lysine) acetyltransferase 2A |
| chr5_+_179125368 | 0.20 |
ENST00000502296.1 ENST00000504734.1 ENST00000415618.2 |
CANX |
calnexin |
| chr22_-_32058166 | 0.20 |
ENST00000435900.1 ENST00000336566.4 |
PISD |
phosphatidylserine decarboxylase |
| chr16_-_28857677 | 0.20 |
ENST00000313511.3 |
TUFM |
Tu translation elongation factor, mitochondrial |
| chr3_-_130745571 | 0.18 |
ENST00000514044.1 ENST00000264992.3 |
ASTE1 |
asteroid homolog 1 (Drosophila) |
| chr7_+_92158083 | 0.18 |
ENST00000265732.5 ENST00000481551.1 ENST00000496410.1 |
RBM48 |
RNA binding motif protein 48 |
| chr8_-_13134045 | 0.18 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr1_+_150980989 | 0.18 |
ENST00000368935.1 |
PRUNE |
prune exopolyphosphatase |
| chr1_+_165864821 | 0.18 |
ENST00000470820.1 |
UCK2 |
uridine-cytidine kinase 2 |
| chr15_-_101835110 | 0.18 |
ENST00000560496.1 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr3_+_138067314 | 0.18 |
ENST00000423968.2 |
MRAS |
muscle RAS oncogene homolog |
| chr3_-_33138286 | 0.18 |
ENST00000416695.2 ENST00000342462.4 ENST00000399402.3 |
TMPPE GLB1 |
transmembrane protein with metallophosphoesterase domain galactosidase, beta 1 |
| chr3_+_138066539 | 0.17 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr8_+_110552046 | 0.17 |
ENST00000529931.1 |
EBAG9 |
estrogen receptor binding site associated, antigen, 9 |
| chr19_+_53073526 | 0.17 |
ENST00000596514.1 ENST00000391785.3 ENST00000301093.2 |
ZNF701 |
zinc finger protein 701 |
| chr5_-_132113083 | 0.17 |
ENST00000296873.7 |
SEPT8 |
septin 8 |
| chr10_-_50970322 | 0.17 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr21_-_26979786 | 0.17 |
ENST00000419219.1 ENST00000352957.4 ENST00000307301.7 |
MRPL39 |
mitochondrial ribosomal protein L39 |
| chr4_-_38666430 | 0.16 |
ENST00000436901.1 |
AC021860.1 |
Uncharacterized protein |
| chr19_-_16582815 | 0.16 |
ENST00000455140.2 ENST00000248070.6 ENST00000594975.1 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
| chr19_-_12267524 | 0.16 |
ENST00000455799.1 ENST00000355738.1 ENST00000439556.2 ENST00000542938.1 |
ZNF625 |
zinc finger protein 625 |
| chr9_+_72873837 | 0.16 |
ENST00000361138.5 |
SMC5 |
structural maintenance of chromosomes 5 |
| chr9_-_80437915 | 0.15 |
ENST00000397476.3 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
| chr11_+_61722629 | 0.15 |
ENST00000526988.1 |
BEST1 |
bestrophin 1 |
| chr10_+_45869652 | 0.14 |
ENST00000542434.1 ENST00000374391.2 |
ALOX5 |
arachidonate 5-lipoxygenase |
| chr17_+_66539369 | 0.14 |
ENST00000600820.1 |
AC079210.1 |
Uncharacterized protein; cDNA FLJ45097 fis, clone BRAWH3031054 |
| chr6_+_37321748 | 0.14 |
ENST00000373479.4 ENST00000394443.4 |
RNF8 |
ring finger protein 8, E3 ubiquitin protein ligase |
| chr11_-_57298187 | 0.14 |
ENST00000525158.1 ENST00000257245.4 ENST00000525587.1 |
TIMM10 |
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr1_+_165864800 | 0.14 |
ENST00000469256.2 |
UCK2 |
uridine-cytidine kinase 2 |
| chr1_+_6640108 | 0.14 |
ENST00000377674.4 ENST00000488936.1 |
ZBTB48 |
zinc finger and BTB domain containing 48 |
| chr1_+_241695670 | 0.13 |
ENST00000366557.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr10_-_50970382 | 0.13 |
ENST00000419399.1 ENST00000432695.1 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr19_+_36120009 | 0.13 |
ENST00000589871.1 |
RBM42 |
RNA binding motif protein 42 |
| chr1_+_93297622 | 0.13 |
ENST00000315741.5 |
RPL5 |
ribosomal protein L5 |
| chr17_-_38074859 | 0.12 |
ENST00000520542.1 ENST00000418519.1 ENST00000394179.1 |
GSDMB |
gasdermin B |
| chr6_-_90529418 | 0.12 |
ENST00000439638.1 ENST00000369393.3 ENST00000428876.1 |
MDN1 |
MDN1, midasin homolog (yeast) |
| chr11_+_92085262 | 0.12 |
ENST00000298047.6 ENST00000409404.2 ENST00000541502.1 |
FAT3 |
FAT atypical cadherin 3 |
| chr2_-_42588338 | 0.12 |
ENST00000234301.2 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
| chr11_+_64009072 | 0.12 |
ENST00000535135.1 ENST00000394540.3 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr11_+_28131821 | 0.12 |
ENST00000379199.2 ENST00000303459.6 |
METTL15 |
methyltransferase like 15 |
| chr19_+_36119975 | 0.12 |
ENST00000589559.1 ENST00000360475.4 |
RBM42 |
RNA binding motif protein 42 |
| chr20_+_56964253 | 0.12 |
ENST00000395802.3 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr4_-_69215467 | 0.11 |
ENST00000579690.1 |
YTHDC1 |
YTH domain containing 1 |
| chr16_-_790982 | 0.11 |
ENST00000301694.5 ENST00000251588.2 |
NARFL |
nuclear prelamin A recognition factor-like |
| chr5_-_132113036 | 0.11 |
ENST00000378706.1 |
SEPT8 |
septin 8 |
| chr2_+_28618532 | 0.11 |
ENST00000545753.1 |
FOSL2 |
FOS-like antigen 2 |
| chr2_-_225434538 | 0.11 |
ENST00000409096.1 |
CUL3 |
cullin 3 |
| chr4_+_128886532 | 0.11 |
ENST00000444616.1 ENST00000388795.5 |
C4orf29 |
chromosome 4 open reading frame 29 |
| chr2_+_89999259 | 0.11 |
ENST00000558026.1 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr17_-_48546232 | 0.10 |
ENST00000258969.4 |
CHAD |
chondroadherin |
| chr17_-_48546324 | 0.10 |
ENST00000508540.1 |
CHAD |
chondroadherin |
| chr5_+_145826867 | 0.10 |
ENST00000296702.5 ENST00000394421.2 |
TCERG1 |
transcription elongation regulator 1 |
| chr15_-_74726283 | 0.10 |
ENST00000543145.2 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
| chr9_+_131266963 | 0.10 |
ENST00000309971.4 ENST00000372770.4 |
GLE1 |
GLE1 RNA export mediator |
| chrX_+_118602363 | 0.10 |
ENST00000317881.8 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
| chr1_+_10459111 | 0.10 |
ENST00000541529.1 ENST00000270776.8 ENST00000483936.1 ENST00000538557.1 |
PGD |
phosphogluconate dehydrogenase |
| chr1_+_179050512 | 0.10 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr7_+_99775520 | 0.10 |
ENST00000317296.5 ENST00000422690.1 ENST00000439782.1 |
STAG3 |
stromal antigen 3 |
| chr8_-_33457453 | 0.10 |
ENST00000523956.1 ENST00000256261.4 |
DUSP26 |
dual specificity phosphatase 26 (putative) |
| chr3_-_15106747 | 0.10 |
ENST00000449354.2 ENST00000444840.2 ENST00000253686.2 |
MRPS25 |
mitochondrial ribosomal protein S25 |
| chr9_-_130890662 | 0.10 |
ENST00000277462.5 ENST00000338961.6 |
PTGES2 |
prostaglandin E synthase 2 |
| chr5_+_177433973 | 0.10 |
ENST00000507848.1 |
FAM153C |
family with sequence similarity 153, member C |
| chr7_+_47834880 | 0.09 |
ENST00000258776.4 |
C7orf69 |
chromosome 7 open reading frame 69 |
| chr12_-_58240470 | 0.09 |
ENST00000548823.1 ENST00000398073.2 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr19_+_10541462 | 0.09 |
ENST00000293683.5 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr7_-_25164868 | 0.09 |
ENST00000409409.1 ENST00000409764.1 ENST00000413447.1 |
CYCS |
cytochrome c, somatic |
| chr7_-_102158157 | 0.08 |
ENST00000541662.1 ENST00000306682.6 ENST00000465829.1 |
RASA4B |
RAS p21 protein activator 4B |
| chr1_+_171454639 | 0.08 |
ENST00000392078.3 ENST00000426496.2 |
PRRC2C |
proline-rich coiled-coil 2C |
| chr11_-_82997371 | 0.08 |
ENST00000525503.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr20_+_57209828 | 0.07 |
ENST00000598340.1 |
MGC4294 |
HCG1785561; MGC4294 protein; Uncharacterized protein |
| chr16_+_50300427 | 0.07 |
ENST00000394697.2 ENST00000566433.2 ENST00000538642.1 |
ADCY7 |
adenylate cyclase 7 |
| chr1_-_160832642 | 0.07 |
ENST00000368034.4 |
CD244 |
CD244 molecule, natural killer cell receptor 2B4 |
| chr1_+_241695424 | 0.07 |
ENST00000366558.3 ENST00000366559.4 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr10_+_104474207 | 0.07 |
ENST00000602831.1 ENST00000369893.5 |
SFXN2 |
sideroflexin 2 |
| chr22_+_18121356 | 0.07 |
ENST00000317582.5 ENST00000543133.1 ENST00000538149.1 ENST00000337612.5 ENST00000493680.1 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
| chr9_-_127533582 | 0.07 |
ENST00000416460.2 |
NR6A1 |
nuclear receptor subfamily 6, group A, member 1 |
| chr15_+_81489213 | 0.07 |
ENST00000559383.1 ENST00000394660.2 |
IL16 |
interleukin 16 |
| chr5_+_64859066 | 0.07 |
ENST00000261308.5 ENST00000535264.1 ENST00000538977.1 |
PPWD1 |
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr19_+_6361754 | 0.07 |
ENST00000597326.1 |
CLPP |
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr2_+_101179152 | 0.06 |
ENST00000264254.6 |
PDCL3 |
phosducin-like 3 |
| chr8_-_37824442 | 0.06 |
ENST00000345060.3 |
ADRB3 |
adrenoceptor beta 3 |
| chr19_+_36119929 | 0.06 |
ENST00000588161.1 ENST00000262633.4 ENST00000592202.1 ENST00000586618.1 |
RBM42 |
RNA binding motif protein 42 |
| chr15_-_41047421 | 0.06 |
ENST00000560460.1 ENST00000338376.3 ENST00000560905.1 |
RMDN3 |
regulator of microtubule dynamics 3 |
| chr17_+_16593539 | 0.06 |
ENST00000340621.5 ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A |
coiled-coil domain containing 144A |
| chr9_-_86571628 | 0.05 |
ENST00000376344.3 |
C9orf64 |
chromosome 9 open reading frame 64 |
| chr2_+_128403439 | 0.05 |
ENST00000544369.1 |
GPR17 |
G protein-coupled receptor 17 |
| chr1_-_44497118 | 0.05 |
ENST00000537678.1 ENST00000466926.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr10_+_60028818 | 0.05 |
ENST00000333926.5 |
CISD1 |
CDGSH iron sulfur domain 1 |
| chr19_+_15752088 | 0.05 |
ENST00000585846.1 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
| chr17_+_21730180 | 0.05 |
ENST00000584398.1 |
UBBP4 |
ubiquitin B pseudogene 4 |
| chr3_-_197024965 | 0.04 |
ENST00000392382.2 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr12_+_121078355 | 0.04 |
ENST00000316803.3 |
CABP1 |
calcium binding protein 1 |
| chr16_-_49890016 | 0.04 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr4_-_69215699 | 0.04 |
ENST00000510746.1 ENST00000344157.4 ENST00000355665.3 |
YTHDC1 |
YTH domain containing 1 |
| chr1_+_93297582 | 0.04 |
ENST00000370321.3 |
RPL5 |
ribosomal protein L5 |
| chr6_-_31628512 | 0.04 |
ENST00000375911.1 |
C6orf47 |
chromosome 6 open reading frame 47 |
| chr1_-_44497024 | 0.04 |
ENST00000372306.3 ENST00000372310.3 ENST00000475075.2 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr6_-_137540477 | 0.03 |
ENST00000367735.2 ENST00000367739.4 ENST00000458076.1 ENST00000414770.1 |
IFNGR1 |
interferon gamma receptor 1 |
| chr10_-_96122682 | 0.03 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr19_-_40732594 | 0.03 |
ENST00000430325.2 ENST00000433940.1 |
CNTD2 |
cyclin N-terminal domain containing 2 |
| chr2_-_136288113 | 0.03 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr19_-_19754354 | 0.02 |
ENST00000587238.1 |
GMIP |
GEM interacting protein |
| chr5_-_177210399 | 0.02 |
ENST00000510276.1 |
FAM153A |
family with sequence similarity 153, member A |
| chr1_-_207226313 | 0.02 |
ENST00000367084.1 |
YOD1 |
YOD1 deubiquitinase |
| chr6_-_8102279 | 0.02 |
ENST00000488226.2 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
| chr16_+_85832146 | 0.02 |
ENST00000565078.1 |
COX4I1 |
cytochrome c oxidase subunit IV isoform 1 |
| chr12_+_20522179 | 0.02 |
ENST00000359062.3 |
PDE3A |
phosphodiesterase 3A, cGMP-inhibited |
| chr19_+_49535169 | 0.02 |
ENST00000474913.1 ENST00000359342.6 |
CGB2 |
chorionic gonadotropin, beta polypeptide 2 |
| chrX_+_16668278 | 0.02 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr16_-_790887 | 0.01 |
ENST00000540986.1 |
NARFL |
nuclear prelamin A recognition factor-like |
| chr11_-_4719072 | 0.01 |
ENST00000396950.3 ENST00000532598.1 |
OR51E2 |
olfactory receptor, family 51, subfamily E, member 2 |
| chr19_+_45754505 | 0.01 |
ENST00000262891.4 ENST00000300843.4 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
| chr15_+_59279851 | 0.01 |
ENST00000348370.4 ENST00000434298.1 ENST00000559160.1 |
RNF111 |
ring finger protein 111 |
| chr5_+_180415839 | 0.01 |
ENST00000342868.6 |
BTNL3 |
butyrophilin-like 3 |
| chr2_-_89247338 | 0.01 |
ENST00000496168.1 |
IGKV1-5 |
immunoglobulin kappa variable 1-5 |
| chr4_+_128886424 | 0.01 |
ENST00000398965.1 |
C4orf29 |
chromosome 4 open reading frame 29 |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 0.5 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.2 | 0.5 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.2 | 0.5 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.5 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.4 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.2 | GO:0031177 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.3 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.1 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.5 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.6 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 2.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.2 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.0 | 0.3 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.3 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.0 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.4 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.1 | 0.4 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.3 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.3 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.1 | 0.4 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.1 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.3 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.5 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.5 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0009368 | endopeptidase Clp complex(GO:0009368) |
| 0.0 | 0.4 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.3 | 0.9 | GO:0090427 | activation of meiosis(GO:0090427) |
| 0.3 | 1.0 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 0.6 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.4 | GO:1905064 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) phenotypic switching(GO:0036166) negative regulation of phospholipid biosynthetic process(GO:0071072) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) regulation of phenotypic switching(GO:1900239) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.1 | 0.5 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.1 | 0.5 | GO:1900738 | cytolysis in other organism involved in symbiotic interaction(GO:0051801) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.1 | 1.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 0.3 | GO:0009224 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.1 | 0.4 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.1 | 0.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.3 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.1 | 0.5 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.5 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.1 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 | 0.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.6 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.1 | 0.3 | GO:0045399 | response to molecule of fungal origin(GO:0002238) regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) cellular response to molecule of fungal origin(GO:0071226) |
| 0.1 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.1 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 1.1 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.1 | 0.4 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.2 | GO:0034184 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.1 | 0.2 | GO:1900154 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.3 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.4 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.2 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.3 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.4 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.1 | GO:0061537 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:2000566 | positive regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000566) |
| 0.0 | 0.4 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0044597 | polyketide metabolic process(GO:0030638) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.3 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:1901029 | adenine transport(GO:0015853) negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0046116 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.4 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.6 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.4 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.0 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.2 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |