Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for RAD21_SMC3
Z-value: 0.90
Transcription factors associated with RAD21_SMC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
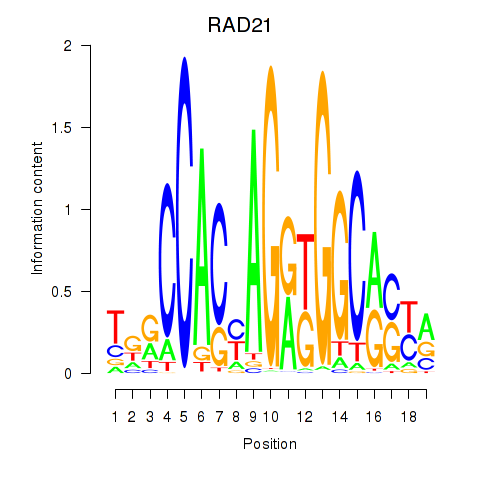
RAD21
|
ENSG00000164754.8 | RAD21 |
|
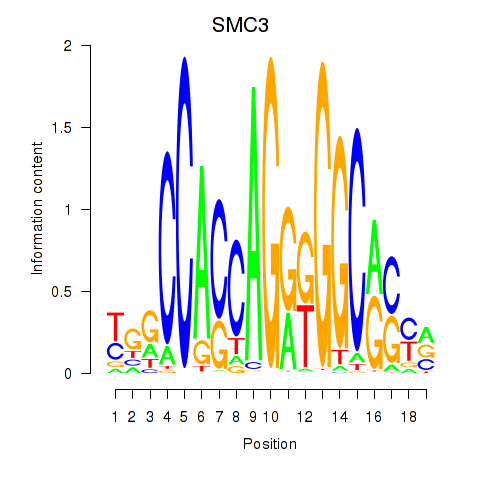
SMC3
|
ENSG00000108055.9 | SMC3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SMC3 | hg19_v2_chr10_+_112327425_112327516 | -0.37 | 1.5e-01 | Click! |
| RAD21 | hg19_v2_chr8_-_117886955_117887105 | -0.03 | 9.2e-01 | Click! |
Activity profile of RAD21_SMC3 motif
Sorted Z-values of RAD21_SMC3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RAD21_SMC3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_17272608 | 3.45 |
ENST00000421459.2 |
VIM |
vimentin |
| chr19_-_55653259 | 2.31 |
ENST00000593194.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr19_-_55652290 | 2.29 |
ENST00000589745.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr15_-_58358607 | 2.12 |
ENST00000249750.4 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr22_-_22901477 | 2.10 |
ENST00000420709.1 ENST00000398741.1 ENST00000405655.3 |
PRAME |
preferentially expressed antigen in melanoma |
| chr22_-_22901636 | 2.07 |
ENST00000406503.1 ENST00000439106.1 ENST00000402697.1 ENST00000543184.1 ENST00000398743.2 |
PRAME |
preferentially expressed antigen in melanoma |
| chr5_-_136834982 | 1.99 |
ENST00000510689.1 ENST00000394945.1 |
SPOCK1 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr7_-_22539771 | 1.55 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr22_-_22292934 | 1.52 |
ENST00000538191.1 ENST00000424647.1 ENST00000407142.1 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr16_+_2867228 | 1.27 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr2_+_69240511 | 1.24 |
ENST00000409349.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr10_-_49732281 | 1.23 |
ENST00000374170.1 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr16_+_2867164 | 1.22 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr1_+_145524891 | 1.21 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr2_+_69240415 | 1.20 |
ENST00000409829.3 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr12_-_51663728 | 1.19 |
ENST00000603864.1 ENST00000605426.1 |
SMAGP |
small cell adhesion glycoprotein |
| chr15_+_63340734 | 1.18 |
ENST00000560959.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr2_+_69240302 | 1.18 |
ENST00000303714.4 |
ANTXR1 |
anthrax toxin receptor 1 |
| chr14_-_75079026 | 1.16 |
ENST00000261978.4 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
| chr1_-_170043709 | 1.13 |
ENST00000367767.1 ENST00000361580.2 ENST00000538366.1 |
KIFAP3 |
kinesin-associated protein 3 |
| chr11_-_407103 | 1.13 |
ENST00000526395.1 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr15_+_63340553 | 1.09 |
ENST00000334895.5 |
TPM1 |
tropomyosin 1 (alpha) |
| chr1_-_6614565 | 1.09 |
ENST00000377705.5 |
NOL9 |
nucleolar protein 9 |
| chr12_-_51663959 | 1.08 |
ENST00000604188.1 ENST00000398453.3 |
SMAGP |
small cell adhesion glycoprotein |
| chr11_-_130786400 | 1.07 |
ENST00000265909.4 |
SNX19 |
sorting nexin 19 |
| chr15_+_63340775 | 1.07 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
| chr1_+_10490779 | 1.05 |
ENST00000477755.1 |
APITD1 |
apoptosis-inducing, TAF9-like domain 1 |
| chr8_-_90996837 | 1.03 |
ENST00000519426.1 ENST00000265433.3 |
NBN |
nibrin |
| chr13_-_88323218 | 1.03 |
ENST00000436290.2 ENST00000453832.2 ENST00000606590.1 |
MIR4500HG |
MIR4500 host gene (non-protein coding) |
| chr3_-_128212016 | 1.02 |
ENST00000498200.1 ENST00000341105.2 |
GATA2 |
GATA binding protein 2 |
| chr22_-_22307199 | 0.99 |
ENST00000397495.4 ENST00000263212.5 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr16_-_19897455 | 0.93 |
ENST00000568214.1 ENST00000569479.1 |
GPRC5B |
G protein-coupled receptor, family C, group 5, member B |
| chr12_+_111843749 | 0.91 |
ENST00000341259.2 |
SH2B3 |
SH2B adaptor protein 3 |
| chr16_-_2390704 | 0.90 |
ENST00000301732.5 ENST00000382381.3 |
ABCA3 |
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chr7_-_27219849 | 0.88 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr1_-_94374946 | 0.87 |
ENST00000370238.3 |
GCLM |
glutamate-cysteine ligase, modifier subunit |
| chr16_+_230435 | 0.85 |
ENST00000199708.2 |
HBQ1 |
hemoglobin, theta 1 |
| chr8_-_90996459 | 0.85 |
ENST00000517337.1 ENST00000409330.1 |
NBN |
nibrin |
| chr1_-_113249734 | 0.84 |
ENST00000484054.3 ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC |
ras homolog family member C |
| chr22_+_19705928 | 0.83 |
ENST00000383045.3 ENST00000438754.2 |
SEPT5 |
septin 5 |
| chr1_-_113249678 | 0.83 |
ENST00000369633.2 ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC |
ras homolog family member C |
| chr1_+_186344883 | 0.81 |
ENST00000367470.3 |
C1orf27 |
chromosome 1 open reading frame 27 |
| chr11_-_113644491 | 0.80 |
ENST00000200135.3 |
ZW10 |
zw10 kinetochore protein |
| chr19_+_48216600 | 0.76 |
ENST00000263277.3 ENST00000538399.1 |
EHD2 |
EH-domain containing 2 |
| chr1_+_186344945 | 0.76 |
ENST00000419367.3 ENST00000287859.6 |
C1orf27 |
chromosome 1 open reading frame 27 |
| chr1_+_193091080 | 0.76 |
ENST00000367435.3 |
CDC73 |
cell division cycle 73 |
| chr10_-_76868931 | 0.75 |
ENST00000372700.3 ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13 |
dual specificity phosphatase 13 |
| chr1_+_151129135 | 0.75 |
ENST00000602841.1 |
SCNM1 |
sodium channel modifier 1 |
| chr3_+_58291965 | 0.73 |
ENST00000445193.3 ENST00000295959.5 ENST00000466547.1 |
RPP14 |
ribonuclease P/MRP 14kDa subunit |
| chr19_+_41313017 | 0.69 |
ENST00000595621.1 ENST00000595051.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr15_+_63340647 | 0.69 |
ENST00000404484.4 |
TPM1 |
tropomyosin 1 (alpha) |
| chr8_-_77912431 | 0.66 |
ENST00000357039.4 ENST00000522527.1 |
PEX2 |
peroxisomal biogenesis factor 2 |
| chr1_+_95699704 | 0.65 |
ENST00000370202.4 |
RWDD3 |
RWD domain containing 3 |
| chr10_+_35416223 | 0.65 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr7_-_5569588 | 0.63 |
ENST00000417101.1 |
ACTB |
actin, beta |
| chr2_+_231921574 | 0.62 |
ENST00000308696.6 ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr12_+_58176525 | 0.61 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr11_-_61647935 | 0.61 |
ENST00000531956.1 |
FADS3 |
fatty acid desaturase 3 |
| chr12_+_123237321 | 0.60 |
ENST00000280557.6 ENST00000455982.2 |
DENR |
density-regulated protein |
| chr16_-_30032610 | 0.60 |
ENST00000574405.1 |
DOC2A |
double C2-like domains, alpha |
| chr6_+_127588020 | 0.60 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr9_+_91003271 | 0.60 |
ENST00000375859.3 ENST00000541629.1 |
SPIN1 |
spindlin 1 |
| chr9_-_94712434 | 0.58 |
ENST00000375708.3 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
| chr9_+_130965677 | 0.54 |
ENST00000393594.3 ENST00000486160.1 |
DNM1 |
dynamin 1 |
| chr9_+_130965651 | 0.54 |
ENST00000475805.1 ENST00000341179.7 ENST00000372923.3 |
DNM1 |
dynamin 1 |
| chr10_-_27149792 | 0.54 |
ENST00000376140.3 ENST00000376170.4 |
ABI1 |
abl-interactor 1 |
| chr1_-_36615051 | 0.54 |
ENST00000373163.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chrX_+_110924346 | 0.54 |
ENST00000371979.3 ENST00000251943.4 ENST00000486353.1 ENST00000394780.3 ENST00000495283.1 |
ALG13 |
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr19_+_49496705 | 0.53 |
ENST00000595090.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr1_-_36615065 | 0.53 |
ENST00000373166.3 ENST00000373159.1 ENST00000373162.1 |
TRAPPC3 |
trafficking protein particle complex 3 |
| chr19_+_49496782 | 0.52 |
ENST00000601968.1 ENST00000596837.1 |
RUVBL2 |
RuvB-like AAA ATPase 2 |
| chr1_+_203830703 | 0.52 |
ENST00000414487.2 |
SNRPE |
small nuclear ribonucleoprotein polypeptide E |
| chr3_-_183735731 | 0.52 |
ENST00000334444.6 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr1_+_21835858 | 0.52 |
ENST00000539907.1 ENST00000540617.1 ENST00000374840.3 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
| chr16_+_2255841 | 0.52 |
ENST00000301725.7 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr9_-_139940608 | 0.52 |
ENST00000371601.4 |
NPDC1 |
neural proliferation, differentiation and control, 1 |
| chr11_-_36531774 | 0.51 |
ENST00000348124.5 ENST00000526995.1 |
TRAF6 |
TNF receptor-associated factor 6, E3 ubiquitin protein ligase |
| chr10_-_6622258 | 0.49 |
ENST00000263125.5 |
PRKCQ |
protein kinase C, theta |
| chr16_-_30441293 | 0.48 |
ENST00000565758.1 ENST00000567983.1 ENST00000319285.4 |
DCTPP1 |
dCTP pyrophosphatase 1 |
| chrX_+_149009941 | 0.48 |
ENST00000535454.1 ENST00000542674.1 ENST00000286482.1 |
MAGEA8 |
melanoma antigen family A, 8 |
| chrX_+_11776701 | 0.48 |
ENST00000476743.1 ENST00000421368.2 ENST00000398527.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr19_+_45842445 | 0.48 |
ENST00000598357.1 |
L47234.1 |
Uncharacterized protein |
| chr10_-_6622201 | 0.48 |
ENST00000539722.1 ENST00000397176.2 |
PRKCQ |
protein kinase C, theta |
| chr11_-_63536113 | 0.47 |
ENST00000433688.1 ENST00000546282.2 |
C11orf95 RP11-466C23.4 |
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr10_+_13628933 | 0.47 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr6_+_127587755 | 0.47 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr16_+_2255710 | 0.46 |
ENST00000397124.1 ENST00000565250.1 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr5_-_146833485 | 0.46 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr11_-_64510409 | 0.45 |
ENST00000394429.1 ENST00000394428.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_-_22034770 | 0.44 |
ENST00000598381.1 |
ZNF43 |
zinc finger protein 43 |
| chr3_-_64009102 | 0.44 |
ENST00000478185.1 ENST00000482510.1 ENST00000497323.1 ENST00000492933.1 ENST00000295901.4 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr9_+_139560197 | 0.43 |
ENST00000371698.3 |
EGFL7 |
EGF-like-domain, multiple 7 |
| chr10_-_6019984 | 0.43 |
ENST00000525219.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr17_-_79827808 | 0.42 |
ENST00000580685.1 |
ARHGDIA |
Rho GDP dissociation inhibitor (GDI) alpha |
| chr10_-_27149851 | 0.42 |
ENST00000376142.2 ENST00000359188.4 ENST00000376139.2 ENST00000376160.1 |
ABI1 |
abl-interactor 1 |
| chr12_-_56583332 | 0.42 |
ENST00000347471.4 ENST00000267064.4 ENST00000394023.3 |
SMARCC2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr10_+_13628921 | 0.41 |
ENST00000378572.3 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr19_-_41859814 | 0.41 |
ENST00000221930.5 |
TGFB1 |
transforming growth factor, beta 1 |
| chrX_+_11776410 | 0.41 |
ENST00000361672.2 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
| chr19_-_22034809 | 0.41 |
ENST00000594012.1 ENST00000595461.1 ENST00000596899.1 |
ZNF43 |
zinc finger protein 43 |
| chr15_-_34502278 | 0.39 |
ENST00000559515.1 ENST00000256544.3 ENST00000560108.1 ENST00000559462.1 |
KATNBL1 |
katanin p80 subunit B-like 1 |
| chr1_+_10270863 | 0.39 |
ENST00000377093.4 ENST00000263934.6 |
KIF1B |
kinesin family member 1B |
| chr14_+_100259666 | 0.39 |
ENST00000262233.6 ENST00000334192.4 |
EML1 |
echinoderm microtubule associated protein like 1 |
| chr19_-_14682838 | 0.39 |
ENST00000215565.2 |
NDUFB7 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa |
| chr5_+_172571445 | 0.39 |
ENST00000231668.9 ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1 |
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr3_-_64009658 | 0.38 |
ENST00000394431.2 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr9_+_100263912 | 0.38 |
ENST00000259365.4 |
TMOD1 |
tropomodulin 1 |
| chrX_+_77166172 | 0.37 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr17_-_34890732 | 0.37 |
ENST00000268852.9 |
MYO19 |
myosin XIX |
| chr1_+_29063119 | 0.37 |
ENST00000474884.1 ENST00000542507.1 |
YTHDF2 |
YTH domain family, member 2 |
| chr17_-_3749515 | 0.37 |
ENST00000158149.3 ENST00000389005.4 |
C17orf85 |
chromosome 17 open reading frame 85 |
| chr2_-_24307162 | 0.36 |
ENST00000413037.1 ENST00000407482.1 |
TP53I3 |
tumor protein p53 inducible protein 3 |
| chr3_+_5163905 | 0.36 |
ENST00000256496.3 ENST00000419534.2 |
ARL8B |
ADP-ribosylation factor-like 8B |
| chr1_-_113249948 | 0.36 |
ENST00000339083.7 ENST00000369642.3 |
RHOC |
ras homolog family member C |
| chr1_+_10490127 | 0.36 |
ENST00000602787.1 ENST00000602296.1 ENST00000400900.2 |
APITD1 APITD1-CORT |
apoptosis-inducing, TAF9-like domain 1 APITD1-CORT readthrough |
| chr1_+_95699740 | 0.36 |
ENST00000429514.2 ENST00000263893.6 |
RWDD3 |
RWD domain containing 3 |
| chr1_-_172413226 | 0.36 |
ENST00000367728.1 ENST00000258324.1 |
PIGC |
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr1_+_36348790 | 0.36 |
ENST00000373204.4 |
AGO1 |
argonaute RISC catalytic component 1 |
| chr8_+_38758737 | 0.35 |
ENST00000521746.1 ENST00000420274.1 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr13_-_37573432 | 0.35 |
ENST00000413537.2 ENST00000443765.1 ENST00000239891.3 |
ALG5 |
ALG5, dolichyl-phosphate beta-glucosyltransferase |
| chr8_-_74659068 | 0.35 |
ENST00000523558.1 ENST00000521210.1 ENST00000355780.5 ENST00000524104.1 ENST00000521736.1 ENST00000521447.1 ENST00000517542.1 ENST00000521451.1 ENST00000521419.1 ENST00000518502.1 ENST00000524300.1 |
STAU2 |
staufen double-stranded RNA binding protein 2 |
| chr1_-_202129704 | 0.35 |
ENST00000476061.1 ENST00000544762.1 ENST00000467283.1 ENST00000464870.1 ENST00000435759.2 ENST00000486116.1 ENST00000543735.1 ENST00000308986.5 ENST00000477625.1 |
PTPN7 |
protein tyrosine phosphatase, non-receptor type 7 |
| chr17_-_33469299 | 0.35 |
ENST00000586869.1 ENST00000360831.5 ENST00000442241.4 |
NLE1 |
notchless homolog 1 (Drosophila) |
| chr12_-_56583243 | 0.35 |
ENST00000550164.1 |
SMARCC2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 |
| chr1_+_10490441 | 0.34 |
ENST00000470413.2 ENST00000309048.3 |
APITD1-CORT APITD1 |
APITD1-CORT readthrough apoptosis-inducing, TAF9-like domain 1 |
| chr10_-_27149904 | 0.34 |
ENST00000376166.1 ENST00000376138.3 ENST00000355394.4 ENST00000346832.5 ENST00000376134.3 ENST00000376137.4 ENST00000536334.1 ENST00000490841.2 |
ABI1 |
abl-interactor 1 |
| chr5_+_148521381 | 0.34 |
ENST00000504238.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr19_-_19051927 | 0.34 |
ENST00000600077.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr16_-_66864806 | 0.34 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr1_-_172413195 | 0.34 |
ENST00000344529.4 |
PIGC |
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr5_-_176889381 | 0.34 |
ENST00000393563.4 ENST00000512501.1 |
DBN1 |
drebrin 1 |
| chr10_+_35416090 | 0.34 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr1_-_226595741 | 0.34 |
ENST00000366794.5 ENST00000366792.1 ENST00000366791.5 |
PARP1 |
poly (ADP-ribose) polymerase 1 |
| chr5_+_148521046 | 0.34 |
ENST00000326685.7 ENST00000356541.3 ENST00000309868.7 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr10_+_13203543 | 0.33 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr17_-_33446820 | 0.33 |
ENST00000592577.1 ENST00000590016.1 ENST00000345365.6 ENST00000360276.3 ENST00000357906.3 |
RAD51D |
RAD51 paralog D |
| chr11_-_117103208 | 0.33 |
ENST00000320934.3 ENST00000530269.1 |
PCSK7 |
proprotein convertase subtilisin/kexin type 7 |
| chr2_-_128399706 | 0.32 |
ENST00000426981.1 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
| chr17_-_26220366 | 0.32 |
ENST00000460380.2 ENST00000508862.1 ENST00000379102.3 ENST00000582441.1 |
LYRM9 RP1-66C13.4 |
LYR motif containing 9 Uncharacterized protein |
| chr10_+_31610064 | 0.32 |
ENST00000446923.2 ENST00000559476.1 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr5_+_34656569 | 0.32 |
ENST00000428746.2 |
RAI14 |
retinoic acid induced 14 |
| chr16_-_67514982 | 0.32 |
ENST00000565835.1 ENST00000540149.1 ENST00000290949.3 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr19_+_24216213 | 0.32 |
ENST00000594934.1 ENST00000597683.1 ENST00000342944.6 |
CTD-2017D11.1 ZNF254 |
CTD-2017D11.1 zinc finger protein 254 |
| chr5_+_34656331 | 0.32 |
ENST00000265109.3 |
RAI14 |
retinoic acid induced 14 |
| chr17_+_16318850 | 0.32 |
ENST00000338560.7 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
| chr17_+_16318909 | 0.31 |
ENST00000577397.1 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
| chr15_-_35261996 | 0.31 |
ENST00000156471.5 |
AQR |
aquarius intron-binding spliceosomal factor |
| chr4_+_71570430 | 0.30 |
ENST00000417478.2 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr3_-_183602515 | 0.30 |
ENST00000449306.1 ENST00000435888.1 ENST00000311101.5 ENST00000317096.4 |
PARL |
presenilin associated, rhomboid-like |
| chr14_-_50319482 | 0.30 |
ENST00000546046.1 ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF |
nuclear export mediator factor |
| chr12_-_96794143 | 0.30 |
ENST00000543119.2 |
CDK17 |
cyclin-dependent kinase 17 |
| chr21_-_34852304 | 0.30 |
ENST00000542230.2 |
TMEM50B |
transmembrane protein 50B |
| chr2_-_24307635 | 0.30 |
ENST00000313482.4 |
TP53I3 |
tumor protein p53 inducible protein 3 |
| chr10_+_35415719 | 0.30 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr1_-_40367668 | 0.29 |
ENST00000397332.2 ENST00000429311.1 |
MYCL |
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chrX_+_153029633 | 0.29 |
ENST00000538966.1 ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3 |
plexin B3 |
| chr17_+_7621045 | 0.29 |
ENST00000570791.1 |
DNAH2 |
dynein, axonemal, heavy chain 2 |
| chr7_-_99679324 | 0.29 |
ENST00000292393.5 ENST00000413658.2 ENST00000412947.1 ENST00000441298.1 ENST00000449785.1 ENST00000299667.4 ENST00000424697.1 |
ZNF3 |
zinc finger protein 3 |
| chr17_+_61562178 | 0.28 |
ENST00000490216.2 |
ACE |
angiotensin I converting enzyme |
| chr5_+_148521136 | 0.28 |
ENST00000506113.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr12_+_104609550 | 0.28 |
ENST00000525566.1 ENST00000429002.2 |
TXNRD1 |
thioredoxin reductase 1 |
| chr3_-_8811288 | 0.27 |
ENST00000316793.3 ENST00000431493.1 |
OXTR |
oxytocin receptor |
| chr10_+_99332529 | 0.27 |
ENST00000455090.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr10_+_99332198 | 0.27 |
ENST00000307518.5 ENST00000298808.5 ENST00000370655.1 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
| chr11_-_64511575 | 0.27 |
ENST00000431822.1 ENST00000377486.3 ENST00000394432.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr17_-_61523622 | 0.27 |
ENST00000448884.2 ENST00000582297.1 ENST00000582034.1 ENST00000578072.1 ENST00000360793.3 |
CYB561 |
cytochrome b561 |
| chr12_+_56618102 | 0.27 |
ENST00000267023.4 ENST00000380198.2 ENST00000341463.5 |
NABP2 |
nucleic acid binding protein 2 |
| chr12_+_51985001 | 0.27 |
ENST00000354534.6 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
| chr17_-_33446735 | 0.26 |
ENST00000460118.2 ENST00000335858.7 |
RAD51D |
RAD51 paralog D |
| chr1_+_154244987 | 0.26 |
ENST00000328703.7 ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1 |
HCLS1 associated protein X-1 |
| chr10_+_35415978 | 0.26 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr14_+_22446680 | 0.26 |
ENST00000390443.3 |
TRAV8-6 |
T cell receptor alpha variable 8-6 |
| chr1_+_29063271 | 0.25 |
ENST00000373812.3 |
YTHDF2 |
YTH domain family, member 2 |
| chr1_+_28844778 | 0.25 |
ENST00000411533.1 |
RCC1 |
regulator of chromosome condensation 1 |
| chr11_+_107879459 | 0.25 |
ENST00000393094.2 |
CUL5 |
cullin 5 |
| chr22_-_32808194 | 0.25 |
ENST00000451746.2 ENST00000216038.5 |
RTCB |
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr12_-_96794330 | 0.25 |
ENST00000261211.3 |
CDK17 |
cyclin-dependent kinase 17 |
| chr3_-_108836977 | 0.25 |
ENST00000232603.5 |
MORC1 |
MORC family CW-type zinc finger 1 |
| chr20_-_30311703 | 0.25 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr5_-_179050066 | 0.25 |
ENST00000329433.6 ENST00000510411.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr12_-_56709786 | 0.25 |
ENST00000547423.1 ENST00000548360.1 ENST00000551475.1 |
RP11-977G19.10 CNPY2 |
Uncharacterized protein canopy FGF signaling regulator 2 |
| chr19_-_49496557 | 0.25 |
ENST00000323798.3 ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1 |
glycogen synthase 1 (muscle) |
| chr7_-_19748640 | 0.25 |
ENST00000222567.5 |
TWISTNB |
TWIST neighbor |
| chr12_-_56709674 | 0.25 |
ENST00000551286.1 ENST00000549318.1 |
CNPY2 RP11-977G19.10 |
canopy FGF signaling regulator 2 Uncharacterized protein |
| chr16_-_47177874 | 0.25 |
ENST00000562435.1 |
NETO2 |
neuropilin (NRP) and tolloid (TLL)-like 2 |
| chr11_+_20409070 | 0.24 |
ENST00000331079.6 |
PRMT3 |
protein arginine methyltransferase 3 |
| chr5_-_146833222 | 0.24 |
ENST00000534907.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr11_-_66206260 | 0.23 |
ENST00000329819.4 ENST00000310999.7 ENST00000430466.2 |
MRPL11 |
mitochondrial ribosomal protein L11 |
| chr9_-_123239632 | 0.23 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr11_+_64948665 | 0.23 |
ENST00000533820.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr17_+_61562201 | 0.23 |
ENST00000290863.6 ENST00000413513.3 ENST00000421982.2 |
ACE |
angiotensin I converting enzyme |
| chr1_+_29063439 | 0.23 |
ENST00000541996.1 ENST00000496288.1 |
YTHDF2 |
YTH domain family, member 2 |
| chr11_+_46368956 | 0.23 |
ENST00000543978.1 |
DGKZ |
diacylglycerol kinase, zeta |
| chr17_+_63133587 | 0.23 |
ENST00000449996.3 ENST00000262406.9 |
RGS9 |
regulator of G-protein signaling 9 |
| chr19_-_49149553 | 0.23 |
ENST00000084798.4 |
CA11 |
carbonic anhydrase XI |
| chr7_-_38407770 | 0.22 |
ENST00000390348.2 |
TRGV1 |
T cell receptor gamma variable 1 (non-functional) |
| chr10_-_96122682 | 0.22 |
ENST00000371361.3 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr5_-_139726181 | 0.22 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr17_-_34890759 | 0.22 |
ENST00000431794.3 |
MYO19 |
myosin XIX |
| chr14_-_102701740 | 0.22 |
ENST00000561150.1 ENST00000522867.1 |
MOK |
MOK protein kinase |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.1 | 1.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.5 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 1.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.0 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 1.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.9 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 2.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.6 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.3 | REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | Genes involved in MyD88:Mal cascade initiated on plasma membrane |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.3 | 0.9 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.3 | 1.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.3 | 1.9 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.3 | 0.8 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 4.0 | GO:0032059 | bleb(GO:0032059) |
| 0.2 | 4.6 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.5 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.9 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 3.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.6 | GO:0033063 | DNA recombinase mediator complex(GO:0033061) Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.1 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.5 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.1 | 2.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 2.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.9 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.1 | 0.7 | GO:0030681 | multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.1 | GO:0097134 | Y chromosome(GO:0000806) cyclin A2-CDK2 complex(GO:0097124) cyclin E1-CDK2 complex(GO:0097134) |
| 0.0 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.6 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.3 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.4 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.6 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 3.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 1.0 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.4 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.0 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 2.5 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.5 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.7 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.3 | 4.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 1.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.3 | 0.9 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 2.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 1.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 1.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.2 | 0.5 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.2 | 2.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.2 | 1.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.4 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.6 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 0.3 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.1 | 0.7 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 4.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 1.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.1 | 2.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.5 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.1 | 0.7 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.3 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.7 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.9 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.1 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.5 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.6 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.3 | GO:0051022 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 1.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.6 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 4.0 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 1.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.9 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 3.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.5 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.0 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 1.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.3 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.2 | GO:0015386 | sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.9 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 1.0 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:1901202 | negative regulation of extracellular matrix assembly(GO:1901202) |
| 0.5 | 4.6 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.5 | 2.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.5 | 4.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.4 | 1.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.4 | 1.1 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 1.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.4 | 1.1 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.3 | 1.0 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.3 | 2.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.3 | 1.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.3 | 3.6 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 4.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.3 | 0.9 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.3 | 2.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.2 | 1.0 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.2 | 1.2 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 2.1 | GO:0035799 | ureter maturation(GO:0035799) |
| 0.2 | 0.9 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.2 | 0.6 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 0.5 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.2 | 1.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.1 | 0.6 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.4 | GO:0052510 | induction by symbiont of host defense response(GO:0044416) induction of host immune response by virus(GO:0046730) active induction of host immune response by virus(GO:0046732) modulation by symbiont of host defense response(GO:0052031) induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) modulation by virus of host immune response(GO:0075528) |
| 0.1 | 0.4 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.5 | GO:1903597 | negative regulation of gap junction assembly(GO:1903597) |
| 0.1 | 0.9 | GO:1903679 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.1 | 0.6 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.3 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.1 | 0.8 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.1 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.5 | GO:0009149 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.1 | 0.8 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.1 | 1.1 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.3 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.3 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.1 | 0.8 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.1 | 1.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.9 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.7 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.1 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.8 | GO:0001711 | endodermal cell fate commitment(GO:0001711) |
| 0.1 | 0.3 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.0 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.0 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.0 | 1.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.7 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 2.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.5 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 2.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.3 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.7 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 1.1 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.0 | 0.5 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.7 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.1 | GO:0072423 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.0 | 0.2 | GO:0045444 | fat cell differentiation(GO:0045444) |
| 0.0 | 0.3 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 0.6 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.4 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 1.9 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:1901889 | negative regulation of cell junction assembly(GO:1901889) |
| 0.0 | 0.3 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0071503 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) |
| 0.0 | 0.4 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.2 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.3 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.0 | 1.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.0 | 0.7 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.0 | GO:0036166 | phenotypic switching(GO:0036166) regulation of phenotypic switching(GO:1900239) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.0 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.0 | 0.1 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.3 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.7 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.4 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) |
| 0.0 | 0.1 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.8 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 1.5 | GO:0051436 | negative regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle(GO:0051436) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 3.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.7 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 2.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.4 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |