Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for RARA
Z-value: 1.04
Transcription factors associated with RARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARA
|
ENSG00000131759.13 | RARA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARA | hg19_v2_chr17_+_38498594_38498661, hg19_v2_chr17_+_38465441_38465481, hg19_v2_chr17_+_38497640_38497647 | -0.32 | 2.3e-01 | Click! |
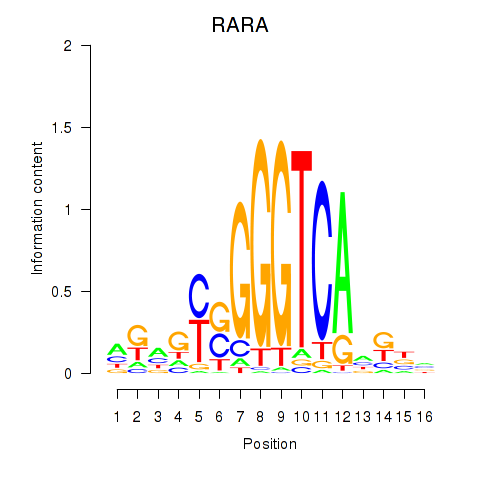
Activity profile of RARA motif
Sorted Z-values of RARA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RARA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_33010175 | 5.09 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr3_-_55521323 | 2.69 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr2_-_151344172 | 2.61 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr1_-_95007193 | 2.37 |
ENST00000370207.4 ENST00000334047.7 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
| chr9_-_21995300 | 2.30 |
ENST00000498628.2 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr10_+_88728189 | 2.28 |
ENST00000416348.1 |
ADIRF |
adipogenesis regulatory factor |
| chr2_-_235405679 | 2.02 |
ENST00000390645.2 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr11_+_10326612 | 1.70 |
ENST00000534464.1 ENST00000530439.1 ENST00000524948.1 ENST00000528655.1 ENST00000526492.1 ENST00000525063.1 |
ADM |
adrenomedullin |
| chr1_-_93426998 | 1.66 |
ENST00000370310.4 |
FAM69A |
family with sequence similarity 69, member A |
| chr9_-_21995249 | 1.63 |
ENST00000494262.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr9_-_21994344 | 1.52 |
ENST00000530628.2 ENST00000361570.3 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr9_+_124030338 | 1.47 |
ENST00000449773.1 ENST00000432226.1 ENST00000436847.1 ENST00000394353.2 ENST00000449733.1 ENST00000412819.1 ENST00000341272.2 ENST00000373808.2 |
GSN |
gelsolin |
| chr9_-_21994597 | 1.47 |
ENST00000579755.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr6_-_46459099 | 1.46 |
ENST00000371374.1 |
RCAN2 |
regulator of calcineurin 2 |
| chr9_+_139873264 | 1.42 |
ENST00000446677.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr13_+_43597269 | 1.38 |
ENST00000379221.2 |
DNAJC15 |
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr1_-_156675368 | 1.38 |
ENST00000368222.3 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr10_+_17270214 | 1.33 |
ENST00000544301.1 |
VIM |
vimentin |
| chr10_-_126849588 | 1.29 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chr8_+_37654693 | 1.24 |
ENST00000412232.2 |
GPR124 |
G protein-coupled receptor 124 |
| chr7_-_45960850 | 1.17 |
ENST00000381083.4 ENST00000381086.5 ENST00000275521.6 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
| chr12_+_54378923 | 1.16 |
ENST00000303460.4 |
HOXC10 |
homeobox C10 |
| chr3_+_45067659 | 1.16 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr1_-_156647189 | 1.12 |
ENST00000368223.3 |
NES |
nestin |
| chr17_-_80291818 | 0.94 |
ENST00000269389.3 ENST00000581691.1 |
SECTM1 |
secreted and transmembrane 1 |
| chr19_+_35168633 | 0.92 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chr19_+_35168567 | 0.92 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chr10_+_43633914 | 0.91 |
ENST00000374466.3 ENST00000374464.1 |
CSGALNACT2 |
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr10_+_89419370 | 0.89 |
ENST00000361175.4 ENST00000456849.1 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chrX_-_37706815 | 0.88 |
ENST00000378578.4 |
DYNLT3 |
dynein, light chain, Tctex-type 3 |
| chr19_+_16435625 | 0.85 |
ENST00000248071.5 ENST00000592003.1 |
KLF2 |
Kruppel-like factor 2 |
| chr8_+_70404996 | 0.83 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chrX_+_51486481 | 0.82 |
ENST00000340438.4 |
GSPT2 |
G1 to S phase transition 2 |
| chr1_-_33168336 | 0.82 |
ENST00000373484.3 |
SYNC |
syncoilin, intermediate filament protein |
| chr19_-_43269809 | 0.79 |
ENST00000406636.3 ENST00000404209.4 ENST00000306511.4 |
PSG8 |
pregnancy specific beta-1-glycoprotein 8 |
| chr9_-_21335356 | 0.75 |
ENST00000359039.4 |
KLHL9 |
kelch-like family member 9 |
| chr6_+_7541845 | 0.73 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr6_-_31324943 | 0.72 |
ENST00000412585.2 ENST00000434333.1 |
HLA-B |
major histocompatibility complex, class I, B |
| chr11_-_8986474 | 0.72 |
ENST00000525069.1 |
TMEM9B |
TMEM9 domain family, member B |
| chr1_-_16939976 | 0.71 |
ENST00000430580.2 |
NBPF1 |
neuroblastoma breakpoint family, member 1 |
| chr1_+_26606608 | 0.70 |
ENST00000319041.6 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr19_-_58662139 | 0.69 |
ENST00000598312.1 |
ZNF329 |
zinc finger protein 329 |
| chr7_-_81399438 | 0.67 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr4_+_154125565 | 0.67 |
ENST00000338700.5 |
TRIM2 |
tripartite motif containing 2 |
| chr5_-_131562935 | 0.66 |
ENST00000379104.2 ENST00000379100.2 ENST00000428369.1 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr9_+_114393634 | 0.66 |
ENST00000556107.1 ENST00000374294.3 |
DNAJC25 DNAJC25-GNG10 |
DnaJ (Hsp40) homolog, subfamily C , member 25 DNAJC25-GNG10 readthrough |
| chr9_-_21335240 | 0.66 |
ENST00000537938.1 |
KLHL9 |
kelch-like family member 9 |
| chr11_-_8985927 | 0.66 |
ENST00000528117.1 ENST00000309134.5 |
TMEM9B |
TMEM9 domain family, member B |
| chr5_-_131563501 | 0.66 |
ENST00000401867.1 ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2 |
prolyl 4-hydroxylase, alpha polypeptide II |
| chr7_-_76247617 | 0.65 |
ENST00000441393.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr16_-_11680791 | 0.65 |
ENST00000571976.1 ENST00000413364.2 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr10_+_115438920 | 0.62 |
ENST00000429617.1 ENST00000369331.4 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr8_-_57358432 | 0.61 |
ENST00000517415.1 ENST00000314922.3 |
PENK |
proenkephalin |
| chr3_+_13590636 | 0.61 |
ENST00000295760.7 |
FBLN2 |
fibulin 2 |
| chr11_-_8986279 | 0.60 |
ENST00000534025.1 |
TMEM9B |
TMEM9 domain family, member B |
| chr8_+_104152922 | 0.60 |
ENST00000309982.5 ENST00000438105.2 ENST00000297574.6 |
BAALC |
brain and acute leukemia, cytoplasmic |
| chr10_+_120967072 | 0.59 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chrX_-_21776281 | 0.58 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr11_-_18610275 | 0.58 |
ENST00000543987.1 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chr19_-_39390440 | 0.57 |
ENST00000249396.7 ENST00000414941.1 ENST00000392081.2 |
SIRT2 |
sirtuin 2 |
| chrX_-_102319092 | 0.56 |
ENST00000372728.3 |
BEX1 |
brain expressed, X-linked 1 |
| chr15_-_65360450 | 0.54 |
ENST00000220062.4 |
RASL12 |
RAS-like, family 12 |
| chr11_+_101983176 | 0.53 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr16_-_11680759 | 0.53 |
ENST00000571459.1 ENST00000570798.1 ENST00000572255.1 ENST00000574763.1 ENST00000574703.1 ENST00000571277.1 ENST00000381810.3 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr22_+_50312379 | 0.53 |
ENST00000407217.3 ENST00000403427.3 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chrX_-_48958043 | 0.52 |
ENST00000356463.3 |
WDR45 |
WD repeat domain 45 |
| chr6_-_57086200 | 0.51 |
ENST00000468148.1 |
RAB23 |
RAB23, member RAS oncogene family |
| chrX_+_54835493 | 0.50 |
ENST00000396224.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chr5_+_131593364 | 0.50 |
ENST00000253754.3 ENST00000379018.3 |
PDLIM4 |
PDZ and LIM domain 4 |
| chr3_+_43732362 | 0.49 |
ENST00000458276.2 |
ABHD5 |
abhydrolase domain containing 5 |
| chr11_-_18610214 | 0.49 |
ENST00000300038.7 ENST00000396197.3 ENST00000320750.6 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chr1_+_43855560 | 0.49 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr20_+_60174827 | 0.49 |
ENST00000543233.1 |
CDH4 |
cadherin 4, type 1, R-cadherin (retinal) |
| chr22_+_50312316 | 0.48 |
ENST00000328268.4 |
CRELD2 |
cysteine-rich with EGF-like domains 2 |
| chr19_-_39390350 | 0.47 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr19_+_8478154 | 0.47 |
ENST00000381035.4 ENST00000595142.1 ENST00000601724.1 ENST00000393944.1 ENST00000215555.2 ENST00000601283.1 ENST00000595213.1 |
MARCH2 |
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr7_-_81399411 | 0.47 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr2_+_85132749 | 0.47 |
ENST00000233143.4 |
TMSB10 |
thymosin beta 10 |
| chr3_-_142608001 | 0.47 |
ENST00000295992.3 |
PCOLCE2 |
procollagen C-endopeptidase enhancer 2 |
| chr20_+_43160409 | 0.46 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr18_+_21529811 | 0.45 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr5_-_114961858 | 0.44 |
ENST00000282382.4 ENST00000456936.3 ENST00000408996.4 |
TMED7-TICAM2 TMED7 TICAM2 |
TMED7-TICAM2 readthrough transmembrane emp24 protein transport domain containing 7 toll-like receptor adaptor molecule 2 |
| chr7_+_143079000 | 0.43 |
ENST00000392910.2 |
ZYX |
zyxin |
| chr2_-_113594279 | 0.43 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr11_-_44971702 | 0.43 |
ENST00000533940.1 ENST00000533937.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr5_-_55290773 | 0.43 |
ENST00000502326.3 ENST00000381298.2 |
IL6ST |
interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| chrX_-_13956737 | 0.42 |
ENST00000454189.2 |
GPM6B |
glycoprotein M6B |
| chr19_+_49458107 | 0.42 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr7_-_105752971 | 0.42 |
ENST00000011473.2 |
SYPL1 |
synaptophysin-like 1 |
| chr11_-_2906979 | 0.42 |
ENST00000380725.1 ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C |
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr11_+_114310237 | 0.41 |
ENST00000539119.1 |
REXO2 |
RNA exonuclease 2 |
| chr12_+_5541267 | 0.40 |
ENST00000423158.3 |
NTF3 |
neurotrophin 3 |
| chr7_+_143078652 | 0.39 |
ENST00000354434.4 ENST00000449423.2 |
ZYX |
zyxin |
| chr22_+_47158578 | 0.39 |
ENST00000355704.3 |
TBC1D22A |
TBC1 domain family, member 22A |
| chr11_+_114310164 | 0.38 |
ENST00000544196.1 ENST00000539754.1 ENST00000539275.1 |
REXO2 |
RNA exonuclease 2 |
| chrX_-_1571810 | 0.38 |
ENST00000381333.4 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr3_+_133292574 | 0.38 |
ENST00000264993.3 |
CDV3 |
CDV3 homolog (mouse) |
| chr11_-_506739 | 0.38 |
ENST00000529306.1 ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr11_+_44117741 | 0.38 |
ENST00000395673.3 ENST00000343631.3 |
EXT2 |
exostosin glycosyltransferase 2 |
| chr11_-_18610246 | 0.38 |
ENST00000379387.4 ENST00000541984.1 |
UEVLD |
UEV and lactate/malate dehyrogenase domains |
| chr22_+_38302285 | 0.37 |
ENST00000215957.6 |
MICALL1 |
MICAL-like 1 |
| chr3_-_142607740 | 0.37 |
ENST00000485766.1 |
PCOLCE2 |
procollagen C-endopeptidase enhancer 2 |
| chr4_+_159593418 | 0.37 |
ENST00000507475.1 ENST00000307738.5 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chrX_+_100878079 | 0.36 |
ENST00000471229.2 |
ARMCX3 |
armadillo repeat containing, X-linked 3 |
| chr9_-_139891165 | 0.36 |
ENST00000494426.1 |
CLIC3 |
chloride intracellular channel 3 |
| chr9_+_21802542 | 0.35 |
ENST00000380172.4 |
MTAP |
methylthioadenosine phosphorylase |
| chr22_+_47158518 | 0.34 |
ENST00000337137.4 ENST00000380995.1 ENST00000407381.3 |
TBC1D22A |
TBC1 domain family, member 22A |
| chr3_+_118905564 | 0.33 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chr19_-_12780211 | 0.33 |
ENST00000597961.1 ENST00000598732.1 ENST00000222190.5 |
CTD-2192J16.24 WDR83OS |
Uncharacterized protein WD repeat domain 83 opposite strand |
| chrX_-_1571759 | 0.32 |
ENST00000381317.3 ENST00000416733.2 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr15_+_80351977 | 0.31 |
ENST00000559157.1 ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr16_+_1359138 | 0.31 |
ENST00000325437.5 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
| chrX_+_55744166 | 0.31 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr11_-_68039364 | 0.31 |
ENST00000533310.1 ENST00000304271.6 ENST00000527280.1 |
C11orf24 |
chromosome 11 open reading frame 24 |
| chrX_+_55744228 | 0.31 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr4_+_159593271 | 0.30 |
ENST00000512251.1 ENST00000511912.1 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr14_+_102027688 | 0.30 |
ENST00000510508.4 ENST00000359323.3 |
DIO3 |
deiodinase, iodothyronine, type III |
| chr1_-_48937821 | 0.30 |
ENST00000396199.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr15_-_34659349 | 0.29 |
ENST00000314891.6 |
LPCAT4 |
lysophosphatidylcholine acyltransferase 4 |
| chr12_+_16035307 | 0.29 |
ENST00000538352.1 ENST00000025399.6 ENST00000419869.2 |
STRAP |
serine/threonine kinase receptor associated protein |
| chr10_-_112678976 | 0.29 |
ENST00000448814.1 |
BBIP1 |
BBSome interacting protein 1 |
| chrX_-_135338503 | 0.28 |
ENST00000370663.5 |
MAP7D3 |
MAP7 domain containing 3 |
| chr12_+_110011571 | 0.28 |
ENST00000539696.1 ENST00000228510.3 ENST00000392727.3 |
MVK |
mevalonate kinase |
| chr17_+_33914276 | 0.28 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr2_+_220283091 | 0.28 |
ENST00000373960.3 |
DES |
desmin |
| chr1_-_43855479 | 0.28 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr17_+_42427826 | 0.28 |
ENST00000586443.1 |
GRN |
granulin |
| chr7_-_105752651 | 0.28 |
ENST00000470347.1 ENST00000455385.2 |
SYPL1 |
synaptophysin-like 1 |
| chr10_-_112678692 | 0.27 |
ENST00000605742.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr1_-_48937838 | 0.27 |
ENST00000371847.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr19_+_16296191 | 0.27 |
ENST00000589852.1 ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A |
family with sequence similarity 32, member A |
| chr1_+_215256467 | 0.27 |
ENST00000391894.2 ENST00000444842.2 |
KCNK2 |
potassium channel, subfamily K, member 2 |
| chr2_+_192542850 | 0.26 |
ENST00000410026.2 |
NABP1 |
nucleic acid binding protein 1 |
| chr19_-_46285646 | 0.26 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr3_+_44596679 | 0.26 |
ENST00000426540.1 ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7 |
zinc finger with KRAB and SCAN domains 7 |
| chr19_-_14228541 | 0.26 |
ENST00000590853.1 ENST00000308677.4 |
PRKACA |
protein kinase, cAMP-dependent, catalytic, alpha |
| chr11_-_33795893 | 0.26 |
ENST00000526785.1 ENST00000534136.1 ENST00000265651.3 ENST00000530401.1 ENST00000448981.2 |
FBXO3 |
F-box protein 3 |
| chrX_-_153640420 | 0.25 |
ENST00000451865.1 ENST00000432135.1 ENST00000369809.1 ENST00000393638.1 ENST00000424626.1 ENST00000309585.5 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr3_+_40428647 | 0.25 |
ENST00000301825.3 ENST00000439533.1 ENST00000456402.1 |
ENTPD3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr9_-_88969303 | 0.25 |
ENST00000277141.6 ENST00000375963.3 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
| chr21_-_46707793 | 0.25 |
ENST00000331343.7 ENST00000349485.5 |
POFUT2 |
protein O-fucosyltransferase 2 |
| chr17_+_33914460 | 0.25 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr3_-_118959733 | 0.25 |
ENST00000459778.1 ENST00000359213.3 |
B4GALT4 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr12_+_6644443 | 0.25 |
ENST00000396858.1 |
GAPDH |
glyceraldehyde-3-phosphate dehydrogenase |
| chr22_-_19279201 | 0.25 |
ENST00000353891.5 ENST00000263200.10 ENST00000427926.1 ENST00000449918.1 |
CLTCL1 |
clathrin, heavy chain-like 1 |
| chr9_-_88969339 | 0.24 |
ENST00000375960.2 ENST00000375961.2 |
ZCCHC6 |
zinc finger, CCHC domain containing 6 |
| chr5_-_146781153 | 0.24 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr5_-_171433579 | 0.24 |
ENST00000265094.5 ENST00000393802.2 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr22_+_45072958 | 0.24 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr10_+_105036909 | 0.24 |
ENST00000369849.4 |
INA |
internexin neuronal intermediate filament protein, alpha |
| chr19_+_39390587 | 0.24 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chrX_+_101975643 | 0.23 |
ENST00000361229.4 |
BHLHB9 |
basic helix-loop-helix domain containing, class B, 9 |
| chr1_+_3816936 | 0.23 |
ENST00000413332.1 ENST00000442673.1 ENST00000439488.1 |
RP13-15E13.1 |
long intergenic non-protein coding RNA 1134 |
| chrX_+_101975619 | 0.23 |
ENST00000457056.1 |
BHLHB9 |
basic helix-loop-helix domain containing, class B, 9 |
| chr1_-_48937682 | 0.22 |
ENST00000371843.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr4_-_186125077 | 0.22 |
ENST00000458385.2 ENST00000514798.1 ENST00000296775.6 |
KIAA1430 |
KIAA1430 |
| chr4_-_2936514 | 0.22 |
ENST00000508221.1 ENST00000507555.1 ENST00000355443.4 |
MFSD10 |
major facilitator superfamily domain containing 10 |
| chr10_-_44144292 | 0.22 |
ENST00000374433.2 |
ZNF32 |
zinc finger protein 32 |
| chr17_+_7210898 | 0.22 |
ENST00000572815.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr1_-_200379180 | 0.22 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr22_+_45072925 | 0.22 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr15_-_63448973 | 0.22 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr1_+_145611010 | 0.22 |
ENST00000369291.5 |
RNF115 |
ring finger protein 115 |
| chr3_-_127317047 | 0.22 |
ENST00000462228.1 ENST00000490643.1 |
TPRA1 |
transmembrane protein, adipocyte asscociated 1 |
| chr10_-_112678904 | 0.21 |
ENST00000423273.1 ENST00000436562.1 ENST00000447005.1 ENST00000454061.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr3_+_133293278 | 0.21 |
ENST00000508481.1 ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3 |
CDV3 homolog (mouse) |
| chr13_-_48575443 | 0.21 |
ENST00000378654.3 |
SUCLA2 |
succinate-CoA ligase, ADP-forming, beta subunit |
| chr3_-_52486841 | 0.20 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr17_-_2304365 | 0.20 |
ENST00000575394.1 ENST00000174618.4 |
MNT |
MAX network transcriptional repressor |
| chr5_+_150827143 | 0.20 |
ENST00000243389.3 ENST00000517945.1 ENST00000521925.1 |
SLC36A1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr14_-_45603657 | 0.19 |
ENST00000396062.3 |
FKBP3 |
FK506 binding protein 3, 25kDa |
| chr17_-_2207062 | 0.19 |
ENST00000263073.6 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr5_-_171433819 | 0.19 |
ENST00000296933.6 |
FBXW11 |
F-box and WD repeat domain containing 11 |
| chr6_-_110500905 | 0.19 |
ENST00000392587.2 |
WASF1 |
WAS protein family, member 1 |
| chrX_+_99899180 | 0.19 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr1_+_109289279 | 0.18 |
ENST00000370008.3 |
STXBP3 |
syntaxin binding protein 3 |
| chr13_-_48575401 | 0.18 |
ENST00000433022.1 ENST00000544100.1 |
SUCLA2 |
succinate-CoA ligase, ADP-forming, beta subunit |
| chr19_-_46285736 | 0.18 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chr10_-_44144152 | 0.18 |
ENST00000395797.1 |
ZNF32 |
zinc finger protein 32 |
| chr11_+_65339820 | 0.17 |
ENST00000316409.2 ENST00000449319.2 ENST00000530349.1 |
FAM89B |
family with sequence similarity 89, member B |
| chr16_+_8891670 | 0.17 |
ENST00000268261.4 ENST00000539622.1 ENST00000569958.1 ENST00000537352.1 |
PMM2 |
phosphomannomutase 2 |
| chr17_+_4843303 | 0.17 |
ENST00000571816.1 |
RNF167 |
ring finger protein 167 |
| chr17_-_2206801 | 0.17 |
ENST00000544865.1 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
| chr4_-_178363581 | 0.17 |
ENST00000264595.2 |
AGA |
aspartylglucosaminidase |
| chr6_-_74233480 | 0.16 |
ENST00000455918.1 |
EEF1A1 |
eukaryotic translation elongation factor 1 alpha 1 |
| chr5_+_150639360 | 0.16 |
ENST00000523004.1 |
GM2A |
GM2 ganglioside activator |
| chr13_+_53029564 | 0.16 |
ENST00000468284.1 ENST00000378034.3 ENST00000258607.5 ENST00000378037.5 |
CKAP2 |
cytoskeleton associated protein 2 |
| chr19_+_58095501 | 0.16 |
ENST00000536878.2 ENST00000597850.1 ENST00000597219.1 ENST00000598689.1 ENST00000599456.1 ENST00000307468.4 |
ZIK1 |
zinc finger protein interacting with K protein 1 |
| chr14_-_64194745 | 0.16 |
ENST00000247225.6 |
SGPP1 |
sphingosine-1-phosphate phosphatase 1 |
| chr10_-_101380121 | 0.16 |
ENST00000370495.4 |
SLC25A28 |
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr6_+_132129151 | 0.16 |
ENST00000360971.2 |
ENPP1 |
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr19_+_18682531 | 0.16 |
ENST00000596304.1 ENST00000430157.2 |
UBA52 |
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr1_-_43855444 | 0.16 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr19_+_54694119 | 0.16 |
ENST00000456872.1 ENST00000302937.4 ENST00000429671.2 |
TSEN34 |
TSEN34 tRNA splicing endonuclease subunit |
| chrX_+_153524024 | 0.15 |
ENST00000369915.3 ENST00000217905.7 |
TKTL1 |
transketolase-like 1 |
| chr17_+_7348658 | 0.15 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr12_+_133613878 | 0.15 |
ENST00000392319.2 ENST00000543758.1 |
ZNF84 |
zinc finger protein 84 |
| chr19_+_39390320 | 0.15 |
ENST00000576510.1 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr19_+_18682661 | 0.15 |
ENST00000596273.1 ENST00000442744.2 ENST00000595683.1 ENST00000599256.1 ENST00000595158.1 ENST00000598780.1 |
UBA52 |
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr4_+_76439665 | 0.15 |
ENST00000508105.1 ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6 |
THAP domain containing 6 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 6.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 0.7 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 2.4 | REACTOME FORMATION OF FIBRIN CLOT CLOTTING CASCADE | Genes involved in Formation of Fibrin Clot (Clotting Cascade) |
| 0.1 | 1.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 4.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 2.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 5.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 3.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.5 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 2.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.4 | 7.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 1.4 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.3 | 1.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.3 | 0.9 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.3 | 1.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.2 | 1.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.3 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.4 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 0.6 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.2 | 0.9 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.2 | 1.2 | GO:0072542 | protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.4 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.4 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) oncostatin-M receptor activity(GO:0004924) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.8 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 2.7 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.4 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.1 | 1.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.5 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 1.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.3 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.7 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.4 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 1.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.1 | 0.2 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.3 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.3 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.4 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 1.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.6 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.2 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.7 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.6 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.3 | 0.8 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 1.2 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 0.8 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.2 | 1.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 1.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 1.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 1.0 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.4 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.6 | GO:0034448 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 1.5 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.6 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.1 | 0.3 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.8 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.9 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.9 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 1.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.8 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) GID complex(GO:0034657) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 2.6 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.0 | 3.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0031305 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 1.6 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.9 | 2.7 | GO:1904933 | cardiac right atrium morphogenesis(GO:0003213) negative regulation of melanin biosynthetic process(GO:0048022) positive regulation of anagen(GO:0051885) mediolateral intercalation(GO:0060031) cervix development(GO:0060067) lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) negative regulation of secondary metabolite biosynthetic process(GO:1900377) regulation of cell proliferation in midbrain(GO:1904933) |
| 0.8 | 2.4 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.6 | 6.6 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.5 | 2.3 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 0.3 | 1.0 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.3 | 1.7 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.3 | 1.5 | GO:1903906 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.3 | 0.8 | GO:0071409 | cellular response to cycloheximide(GO:0071409) |
| 0.3 | 0.8 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.9 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.2 | 0.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.4 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.2 | 1.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 0.9 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.1 | 0.4 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 1.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 1.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.1 | 1.3 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.1 | 0.8 | GO:0060686 | regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.1 | 0.3 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.7 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 0.4 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.5 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 0.7 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.5 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.4 | GO:0046502 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.1 | 0.3 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.3 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.6 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 1.3 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.4 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.3 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.2 | GO:0035606 | induction of programmed cell death(GO:0012502) peptidyl-cysteine S-trans-nitrosylation(GO:0035606) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.1 | 0.6 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.8 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.1 | 0.1 | GO:2000309 | positive regulation of tumor necrosis factor (ligand) superfamily member 11 production(GO:2000309) |
| 0.1 | 0.4 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.1 | 0.7 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 1.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 0.5 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.5 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.6 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.1 | 0.4 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 2.5 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.1 | 0.5 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.1 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 2.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 1.6 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.3 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.2 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.4 | GO:1904354 | negative regulation of telomere capping(GO:1904354) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.3 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.5 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.4 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0045124 | regulation of bone resorption(GO:0045124) |
| 0.0 | 0.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 1.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.8 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.4 | GO:0050930 | induction of positive chemotaxis(GO:0050930) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.3 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.3 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.5 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.0 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.0 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.0 | 0.2 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:2000784 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) regulation of barbed-end actin filament capping(GO:2000812) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.0 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.0 | 0.7 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0043243 | positive regulation of protein complex disassembly(GO:0043243) |
| 0.0 | 0.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |