Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
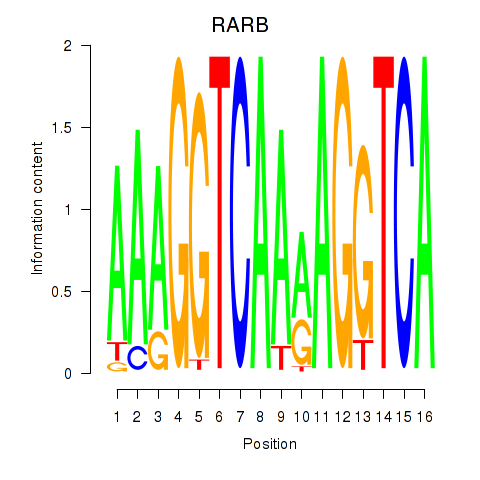
Results for RARB
Z-value: 1.11
Transcription factors associated with RARB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARB
|
ENSG00000077092.14 | RARB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RARB | hg19_v2_chr3_+_25469724_25469773 | -0.13 | 6.3e-01 | Click! |
Activity profile of RARB motif
Sorted Z-values of RARB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RARB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_9731872 | 2.47 |
ENST00000424629.1 ENST00000326044.5 ENST00000354661.4 ENST00000435550.1 ENST00000444611.1 ENST00000421525.1 |
ZNF561 |
zinc finger protein 561 |
| chr7_+_73623717 | 2.31 |
ENST00000344995.5 ENST00000460943.1 |
LAT2 |
linker for activation of T cells family, member 2 |
| chr2_-_21266935 | 2.17 |
ENST00000233242.1 |
APOB |
apolipoprotein B |
| chr6_+_24495067 | 2.05 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr17_-_28618948 | 1.70 |
ENST00000261714.6 |
BLMH |
bleomycin hydrolase |
| chr17_-_62097904 | 1.44 |
ENST00000583366.1 |
ICAM2 |
intercellular adhesion molecule 2 |
| chr7_-_144533074 | 1.42 |
ENST00000360057.3 ENST00000378099.3 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr7_+_73624327 | 1.26 |
ENST00000361082.3 ENST00000275635.7 ENST00000470709.1 |
LAT2 |
linker for activation of T cells family, member 2 |
| chr2_-_175499294 | 1.22 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr17_+_58677539 | 1.18 |
ENST00000305921.3 |
PPM1D |
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr11_-_63381823 | 1.14 |
ENST00000323646.5 |
PLA2G16 |
phospholipase A2, group XVI |
| chr11_-_116708302 | 1.07 |
ENST00000375320.1 ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1 |
apolipoprotein A-I |
| chr11_-_63381925 | 1.06 |
ENST00000415826.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chrX_-_70329118 | 1.04 |
ENST00000374188.3 |
IL2RG |
interleukin 2 receptor, gamma |
| chr1_-_111506562 | 0.95 |
ENST00000485275.2 ENST00000369763.4 |
LRIF1 |
ligand dependent nuclear receptor interacting factor 1 |
| chr11_+_61520075 | 0.94 |
ENST00000278836.5 |
MYRF |
myelin regulatory factor |
| chr15_+_81591757 | 0.92 |
ENST00000558332.1 |
IL16 |
interleukin 16 |
| chr17_-_26903900 | 0.90 |
ENST00000395319.3 ENST00000581807.1 ENST00000584086.1 ENST00000395321.2 |
ALDOC |
aldolase C, fructose-bisphosphate |
| chr8_-_27462822 | 0.90 |
ENST00000522098.1 |
CLU |
clusterin |
| chr1_+_28206150 | 0.87 |
ENST00000456990.1 |
THEMIS2 |
thymocyte selection associated family member 2 |
| chr2_+_68592305 | 0.83 |
ENST00000234313.7 |
PLEK |
pleckstrin |
| chr5_+_125935960 | 0.82 |
ENST00000297540.4 |
PHAX |
phosphorylated adaptor for RNA export |
| chr14_+_95078714 | 0.81 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr19_-_41256207 | 0.75 |
ENST00000598485.2 ENST00000470681.1 ENST00000339153.3 ENST00000598729.1 |
C19orf54 |
chromosome 19 open reading frame 54 |
| chr16_-_1993260 | 0.74 |
ENST00000361871.3 |
MSRB1 |
methionine sulfoxide reductase B1 |
| chr7_-_5998714 | 0.73 |
ENST00000539903.1 |
RSPH10B |
radial spoke head 10 homolog B (Chlamydomonas) |
| chr11_+_308217 | 0.73 |
ENST00000602569.1 |
IFITM2 |
interferon induced transmembrane protein 2 |
| chr10_-_65028938 | 0.70 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr5_-_133968529 | 0.70 |
ENST00000402673.2 |
SAR1B |
SAR1 homolog B (S. cerevisiae) |
| chr13_-_47012325 | 0.69 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr10_-_101825151 | 0.68 |
ENST00000441382.1 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chrX_-_119709637 | 0.66 |
ENST00000404115.3 |
CUL4B |
cullin 4B |
| chr4_-_100140331 | 0.66 |
ENST00000407820.2 ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6 |
alcohol dehydrogenase 6 (class V) |
| chr2_+_88991162 | 0.65 |
ENST00000283646.4 |
RPIA |
ribose 5-phosphate isomerase A |
| chr19_+_49468558 | 0.63 |
ENST00000331825.6 |
FTL |
ferritin, light polypeptide |
| chr4_+_70146217 | 0.62 |
ENST00000335568.5 ENST00000511240.1 |
UGT2B28 |
UDP glucuronosyltransferase 2 family, polypeptide B28 |
| chr2_-_97405775 | 0.62 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr6_-_31763721 | 0.62 |
ENST00000375663.3 |
VARS |
valyl-tRNA synthetase |
| chr11_+_63870660 | 0.61 |
ENST00000246841.3 |
FLRT1 |
fibronectin leucine rich transmembrane protein 1 |
| chr18_+_657578 | 0.61 |
ENST00000323274.10 |
TYMS |
thymidylate synthetase |
| chr1_-_160313025 | 0.61 |
ENST00000368069.3 ENST00000241704.7 |
COPA |
coatomer protein complex, subunit alpha |
| chr4_+_124317940 | 0.61 |
ENST00000505319.1 ENST00000339241.1 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr11_+_95523621 | 0.60 |
ENST00000325542.5 ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57 |
centrosomal protein 57kDa |
| chr6_+_49431073 | 0.59 |
ENST00000335783.3 |
CENPQ |
centromere protein Q |
| chr19_+_50919056 | 0.59 |
ENST00000599632.1 |
CTD-2545M3.6 |
CTD-2545M3.6 |
| chr8_+_74903580 | 0.59 |
ENST00000284818.2 ENST00000518893.1 |
LY96 |
lymphocyte antigen 96 |
| chr1_+_248020481 | 0.57 |
ENST00000366481.3 |
TRIM58 |
tripartite motif containing 58 |
| chr10_-_65028817 | 0.56 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr3_+_127317066 | 0.55 |
ENST00000265056.7 |
MCM2 |
minichromosome maintenance complex component 2 |
| chr1_-_15850676 | 0.52 |
ENST00000440484.1 ENST00000333868.5 |
CASP9 |
caspase 9, apoptosis-related cysteine peptidase |
| chr12_+_57943781 | 0.51 |
ENST00000455537.2 ENST00000286452.5 |
KIF5A |
kinesin family member 5A |
| chr1_+_10534944 | 0.49 |
ENST00000356607.4 ENST00000538836.1 ENST00000491661.2 |
PEX14 |
peroxisomal biogenesis factor 14 |
| chr19_+_21688366 | 0.49 |
ENST00000358491.4 ENST00000597078.1 |
ZNF429 |
zinc finger protein 429 |
| chr22_-_37976082 | 0.46 |
ENST00000215886.4 |
LGALS2 |
lectin, galactoside-binding, soluble, 2 |
| chr3_+_119316689 | 0.46 |
ENST00000273371.4 |
PLA1A |
phospholipase A1 member A |
| chr13_+_73302047 | 0.46 |
ENST00000377814.2 ENST00000377815.3 ENST00000390667.5 |
BORA |
bora, aurora kinase A activator |
| chr9_+_131217459 | 0.44 |
ENST00000497812.2 ENST00000393533.2 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr1_+_17944832 | 0.44 |
ENST00000167825.4 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr16_+_20462783 | 0.43 |
ENST00000574251.1 ENST00000576361.1 ENST00000417235.2 ENST00000573854.1 ENST00000424070.1 ENST00000536134.1 ENST00000219054.6 ENST00000575690.1 ENST00000571894.1 |
ACSM2A |
acyl-CoA synthetase medium-chain family member 2A |
| chr6_-_31763408 | 0.42 |
ENST00000444930.2 |
VARS |
valyl-tRNA synthetase |
| chr1_+_174669653 | 0.42 |
ENST00000325589.5 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr17_+_7155343 | 0.41 |
ENST00000573513.1 ENST00000354429.2 ENST00000574255.1 ENST00000396627.2 ENST00000356683.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr8_+_48873453 | 0.41 |
ENST00000523944.1 |
MCM4 |
minichromosome maintenance complex component 4 |
| chr14_-_39572345 | 0.40 |
ENST00000548032.2 ENST00000556092.1 ENST00000557280.1 ENST00000545328.2 ENST00000553970.1 |
SEC23A |
Sec23 homolog A (S. cerevisiae) |
| chr16_-_20587599 | 0.40 |
ENST00000566384.1 ENST00000565232.1 ENST00000567001.1 ENST00000565322.1 ENST00000569344.1 ENST00000329697.6 ENST00000414188.2 ENST00000568882.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr21_-_35284635 | 0.40 |
ENST00000429238.1 |
AP000304.12 |
AP000304.12 |
| chr10_+_11784360 | 0.39 |
ENST00000379215.4 ENST00000420401.1 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
| chr20_-_32262165 | 0.39 |
ENST00000606690.1 ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
| chr19_+_35849723 | 0.38 |
ENST00000594310.1 |
FFAR3 |
free fatty acid receptor 3 |
| chr6_+_31554962 | 0.37 |
ENST00000376092.3 ENST00000376086.3 ENST00000303757.8 ENST00000376093.2 ENST00000376102.3 |
LST1 |
leukocyte specific transcript 1 |
| chr18_-_7117813 | 0.37 |
ENST00000389658.3 |
LAMA1 |
laminin, alpha 1 |
| chr7_-_74489609 | 0.36 |
ENST00000329959.4 ENST00000503250.2 ENST00000543840.1 |
WBSCR16 |
Williams-Beuren syndrome chromosome region 16 |
| chrX_+_129535937 | 0.34 |
ENST00000305536.6 ENST00000370947.1 |
RBMX2 |
RNA binding motif protein, X-linked 2 |
| chr6_+_24495185 | 0.33 |
ENST00000348925.2 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr7_-_123198284 | 0.33 |
ENST00000355749.2 |
NDUFA5 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr1_+_66258846 | 0.32 |
ENST00000341517.4 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr20_-_3185279 | 0.32 |
ENST00000354488.3 ENST00000380201.2 |
DDRGK1 |
DDRGK domain containing 1 |
| chr3_+_119316721 | 0.31 |
ENST00000488919.1 ENST00000495992.1 |
PLA1A |
phospholipase A1 member A |
| chr22_-_31063782 | 0.30 |
ENST00000404885.1 ENST00000403268.1 ENST00000407308.1 ENST00000342474.4 ENST00000334679.3 |
DUSP18 |
dual specificity phosphatase 18 |
| chr17_+_41363854 | 0.30 |
ENST00000588693.1 ENST00000588659.1 ENST00000541594.1 ENST00000536052.1 ENST00000331615.3 |
TMEM106A |
transmembrane protein 106A |
| chr7_-_6098770 | 0.29 |
ENST00000536084.1 ENST00000446699.1 ENST00000199389.6 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr11_-_506316 | 0.28 |
ENST00000532055.1 ENST00000531540.1 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr6_-_52149475 | 0.28 |
ENST00000419835.2 ENST00000229854.7 ENST00000596288.1 |
MCM3 |
minichromosome maintenance complex component 3 |
| chrX_-_107975917 | 0.28 |
ENST00000563887.1 |
RP6-24A23.6 |
Uncharacterized protein |
| chr11_-_47788847 | 0.27 |
ENST00000263773.5 |
FNBP4 |
formin binding protein 4 |
| chr5_-_147211190 | 0.27 |
ENST00000510027.2 |
SPINK1 |
serine peptidase inhibitor, Kazal type 1 |
| chr19_-_54618650 | 0.27 |
ENST00000391757.1 |
TFPT |
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr11_-_1587166 | 0.26 |
ENST00000331588.4 |
DUSP8 |
dual specificity phosphatase 8 |
| chr12_-_6716569 | 0.26 |
ENST00000544040.1 ENST00000545942.1 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
| chr14_-_24658053 | 0.26 |
ENST00000354464.6 |
IPO4 |
importin 4 |
| chr6_-_49430886 | 0.26 |
ENST00000274813.3 |
MUT |
methylmalonyl CoA mutase |
| chr6_+_31554779 | 0.25 |
ENST00000376090.2 |
LST1 |
leukocyte specific transcript 1 |
| chr12_+_57810198 | 0.25 |
ENST00000598001.1 |
AC126614.1 |
HCG1818482; Uncharacterized protein |
| chr8_-_95274536 | 0.24 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr17_-_41277317 | 0.23 |
ENST00000497488.1 ENST00000489037.1 ENST00000470026.1 ENST00000586385.1 ENST00000591534.1 ENST00000591849.1 |
BRCA1 |
breast cancer 1, early onset |
| chr17_-_41277370 | 0.23 |
ENST00000476777.1 ENST00000491747.2 ENST00000478531.1 ENST00000477152.1 ENST00000357654.3 ENST00000493795.1 ENST00000493919.1 |
BRCA1 |
breast cancer 1, early onset |
| chr5_+_53751445 | 0.23 |
ENST00000302005.1 |
HSPB3 |
heat shock 27kDa protein 3 |
| chr6_-_24667180 | 0.23 |
ENST00000545995.1 |
TDP2 |
tyrosyl-DNA phosphodiesterase 2 |
| chr12_+_6881678 | 0.23 |
ENST00000441671.2 ENST00000203629.2 |
LAG3 |
lymphocyte-activation gene 3 |
| chr11_+_118938485 | 0.23 |
ENST00000300793.6 |
VPS11 |
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr7_+_99425633 | 0.23 |
ENST00000354829.2 ENST00000421837.2 ENST00000417625.1 ENST00000342499.4 ENST00000444905.1 ENST00000415413.1 ENST00000312017.5 ENST00000222382.5 |
CYP3A43 |
cytochrome P450, family 3, subfamily A, polypeptide 43 |
| chr19_-_33360647 | 0.23 |
ENST00000590341.1 ENST00000587772.1 ENST00000023064.4 |
SLC7A9 |
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr6_-_24667232 | 0.23 |
ENST00000378198.4 |
TDP2 |
tyrosyl-DNA phosphodiesterase 2 |
| chr17_+_35767301 | 0.22 |
ENST00000225396.6 ENST00000417170.1 ENST00000590005.1 ENST00000590957.1 |
TADA2A |
transcriptional adaptor 2A |
| chr11_+_31531291 | 0.22 |
ENST00000350638.5 ENST00000379163.5 ENST00000395934.2 |
ELP4 |
elongator acetyltransferase complex subunit 4 |
| chr5_+_446253 | 0.21 |
ENST00000315013.5 |
EXOC3 |
exocyst complex component 3 |
| chr9_-_125590818 | 0.21 |
ENST00000259467.4 |
PDCL |
phosducin-like |
| chr19_+_21324827 | 0.21 |
ENST00000600692.1 ENST00000599296.1 ENST00000594425.1 ENST00000311048.7 |
ZNF431 |
zinc finger protein 431 |
| chr6_+_76311568 | 0.20 |
ENST00000370014.3 |
SENP6 |
SUMO1/sentrin specific peptidase 6 |
| chr4_+_128802016 | 0.20 |
ENST00000270861.5 ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4 |
polo-like kinase 4 |
| chr6_-_35480705 | 0.20 |
ENST00000229771.6 |
TULP1 |
tubby like protein 1 |
| chr17_-_44657017 | 0.20 |
ENST00000573185.1 ENST00000570550.1 ENST00000445552.2 ENST00000336125.5 ENST00000329240.4 ENST00000337845.7 |
ARL17A |
ADP-ribosylation factor-like 17A |
| chr19_+_35862192 | 0.20 |
ENST00000597214.1 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
| chr2_+_108443388 | 0.19 |
ENST00000354986.4 ENST00000408999.3 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
| chr4_-_48082192 | 0.18 |
ENST00000507351.1 |
TXK |
TXK tyrosine kinase |
| chr6_+_42141029 | 0.17 |
ENST00000372958.1 |
GUCA1A |
guanylate cyclase activator 1A (retina) |
| chr18_-_32924372 | 0.16 |
ENST00000261332.6 ENST00000399061.3 |
ZNF24 |
zinc finger protein 24 |
| chr1_+_16330723 | 0.16 |
ENST00000329454.2 |
C1orf64 |
chromosome 1 open reading frame 64 |
| chr19_+_44507091 | 0.15 |
ENST00000429154.2 ENST00000585632.1 |
ZNF230 |
zinc finger protein 230 |
| chr6_+_76311736 | 0.15 |
ENST00000447266.2 |
SENP6 |
SUMO1/sentrin specific peptidase 6 |
| chr11_+_65029233 | 0.15 |
ENST00000265465.3 |
POLA2 |
polymerase (DNA directed), alpha 2, accessory subunit |
| chr16_-_1275257 | 0.15 |
ENST00000234798.4 |
TPSG1 |
tryptase gamma 1 |
| chr19_+_35861831 | 0.14 |
ENST00000454971.1 |
GPR42 |
G protein-coupled receptor 42 (gene/pseudogene) |
| chr3_+_185304059 | 0.14 |
ENST00000427465.2 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr10_-_103599591 | 0.14 |
ENST00000348850.5 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr9_-_100459639 | 0.14 |
ENST00000375128.4 |
XPA |
xeroderma pigmentosum, complementation group A |
| chr1_+_59775752 | 0.14 |
ENST00000371212.1 |
FGGY |
FGGY carbohydrate kinase domain containing |
| chr17_+_28884130 | 0.13 |
ENST00000580161.1 |
TBC1D29 |
TBC1 domain family, member 29 |
| chr4_-_84406218 | 0.13 |
ENST00000515303.1 |
FAM175A |
family with sequence similarity 175, member A |
| chr2_-_107084826 | 0.13 |
ENST00000304514.7 ENST00000409886.3 |
RGPD3 |
RANBP2-like and GRIP domain containing 3 |
| chr6_-_33771757 | 0.12 |
ENST00000507738.1 ENST00000266003.5 ENST00000430124.2 |
MLN |
motilin |
| chr16_+_70380732 | 0.11 |
ENST00000302243.7 |
DDX19A |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
| chr1_-_79472365 | 0.11 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chrX_-_134429952 | 0.11 |
ENST00000370764.1 |
ZNF75D |
zinc finger protein 75D |
| chr15_-_74504597 | 0.11 |
ENST00000416286.3 |
STRA6 |
stimulated by retinoic acid 6 |
| chr16_+_56995854 | 0.11 |
ENST00000566128.1 |
CETP |
cholesteryl ester transfer protein, plasma |
| chr8_+_67624653 | 0.11 |
ENST00000521198.2 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
| chr6_-_28367510 | 0.11 |
ENST00000361028.1 |
ZSCAN12 |
zinc finger and SCAN domain containing 12 |
| chr2_-_176866978 | 0.10 |
ENST00000392540.2 ENST00000409660.1 ENST00000544803.1 ENST00000272748.4 |
KIAA1715 |
KIAA1715 |
| chr9_+_130186653 | 0.10 |
ENST00000342483.5 ENST00000543471.1 |
ZNF79 |
zinc finger protein 79 |
| chr10_-_27389320 | 0.09 |
ENST00000436985.2 |
ANKRD26 |
ankyrin repeat domain 26 |
| chr17_+_26800648 | 0.09 |
ENST00000545060.1 |
SLC13A2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr19_+_39881951 | 0.08 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chr1_+_12834984 | 0.08 |
ENST00000357726.4 |
PRAMEF12 |
PRAME family member 12 |
| chr7_+_20686946 | 0.08 |
ENST00000443026.2 ENST00000406935.1 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr11_+_111782934 | 0.08 |
ENST00000304298.3 |
HSPB2 |
Homo sapiens heat shock 27kDa protein 2 (HSPB2), mRNA. |
| chr11_-_89653576 | 0.08 |
ENST00000420869.1 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chr1_+_221051699 | 0.08 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr22_+_21336267 | 0.08 |
ENST00000215739.8 |
LZTR1 |
leucine-zipper-like transcription regulator 1 |
| chr2_-_152830441 | 0.08 |
ENST00000534999.1 ENST00000397327.2 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chrX_+_73164149 | 0.08 |
ENST00000602938.1 ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX |
JPX transcript, XIST activator (non-protein coding) |
| chr12_-_52761262 | 0.08 |
ENST00000257901.3 |
KRT85 |
keratin 85 |
| chr7_+_73624276 | 0.07 |
ENST00000475494.1 ENST00000398475.1 |
LAT2 |
linker for activation of T cells family, member 2 |
| chr17_+_26800756 | 0.07 |
ENST00000537681.1 |
SLC13A2 |
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr12_-_323689 | 0.07 |
ENST00000428720.1 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr7_-_99381798 | 0.07 |
ENST00000415003.1 ENST00000354593.2 |
CYP3A4 |
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr1_+_78245303 | 0.07 |
ENST00000370791.3 ENST00000443751.2 |
FAM73A |
family with sequence similarity 73, member A |
| chr8_-_141774467 | 0.06 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr17_+_48046538 | 0.05 |
ENST00000240306.3 |
DLX4 |
distal-less homeobox 4 |
| chr12_-_81763184 | 0.05 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr6_+_31637944 | 0.05 |
ENST00000375864.4 |
LY6G5B |
lymphocyte antigen 6 complex, locus G5B |
| chr16_+_2255841 | 0.05 |
ENST00000301725.7 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr1_+_53392901 | 0.05 |
ENST00000371514.3 ENST00000528311.1 ENST00000371509.4 ENST00000407246.2 ENST00000371513.5 |
SCP2 |
sterol carrier protein 2 |
| chr1_-_110950255 | 0.04 |
ENST00000483260.1 ENST00000474861.2 ENST00000602318.1 |
LAMTOR5 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr1_-_110950564 | 0.04 |
ENST00000256644.4 |
LAMTOR5 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr3_+_49297518 | 0.04 |
ENST00000440528.3 |
RP11-3B7.1 |
Uncharacterized protein |
| chr5_+_147774275 | 0.04 |
ENST00000513826.1 |
FBXO38 |
F-box protein 38 |
| chr1_-_23521222 | 0.04 |
ENST00000374619.1 |
HTR1D |
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr3_+_184058125 | 0.04 |
ENST00000310585.4 |
FAM131A |
family with sequence similarity 131, member A |
| chr2_+_11679963 | 0.04 |
ENST00000263834.5 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr13_+_28519343 | 0.03 |
ENST00000381026.3 |
ATP5EP2 |
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit pseudogene 2 |
| chr2_-_136594740 | 0.03 |
ENST00000264162.2 |
LCT |
lactase |
| chr16_-_4852915 | 0.03 |
ENST00000322048.7 |
ROGDI |
rogdi homolog (Drosophila) |
| chr19_+_6361440 | 0.03 |
ENST00000245816.4 |
CLPP |
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr2_+_177025619 | 0.02 |
ENST00000410016.1 |
HOXD3 |
homeobox D3 |
| chr6_+_24667257 | 0.02 |
ENST00000537591.1 ENST00000230048.4 |
ACOT13 |
acyl-CoA thioesterase 13 |
| chr18_-_19283649 | 0.02 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr1_+_9005917 | 0.02 |
ENST00000549778.1 ENST00000480186.3 ENST00000377443.2 ENST00000377436.3 ENST00000377442.2 |
CA6 |
carbonic anhydrase VI |
| chr19_-_23869999 | 0.01 |
ENST00000601935.1 ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675 |
zinc finger protein 675 |
| chr4_+_8594364 | 0.01 |
ENST00000360986.4 |
CPZ |
carboxypeptidase Z |
| chr12_+_104682496 | 0.01 |
ENST00000378070.4 |
TXNRD1 |
thioredoxin reductase 1 |
| chr1_+_52682052 | 0.01 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr1_-_3713042 | 0.01 |
ENST00000378251.1 |
LRRC47 |
leucine rich repeat containing 47 |
| chr8_+_24241969 | 0.01 |
ENST00000522298.1 |
ADAMDEC1 |
ADAM-like, decysin 1 |
| chr1_-_23520755 | 0.01 |
ENST00000314113.3 |
HTR1D |
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr7_-_99381884 | 0.01 |
ENST00000336411.2 |
CYP3A4 |
cytochrome P450, family 3, subfamily A, polypeptide 4 |
| chr2_-_152830479 | 0.01 |
ENST00000360283.6 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
| chr20_-_43729750 | 0.00 |
ENST00000537075.1 ENST00000306117.1 |
KCNS1 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
| chr16_+_2255710 | 0.00 |
ENST00000397124.1 ENST00000565250.1 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0006540 | glutamate decarboxylation to succinate(GO:0006540) gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.4 | 1.4 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.3 | 1.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.3 | 2.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 2.2 | GO:0006642 | triglyceride mobilization(GO:0006642) response to selenium ion(GO:0010269) |
| 0.3 | 1.7 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.3 | 1.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.2 | 0.6 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.2 | 0.8 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.2 | 0.6 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.2 | 1.8 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.2 | 0.5 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.2 | 0.5 | GO:0070512 | regulation of histone H4-K20 methylation(GO:0070510) positive regulation of histone H4-K20 methylation(GO:0070512) |
| 0.2 | 0.6 | GO:0019860 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) transformation of host cell by virus(GO:0019087) uracil metabolic process(GO:0019860) |
| 0.1 | 1.0 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 0.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 0.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 1.0 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 1.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.3 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 0.6 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.4 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 0.6 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.1 | 0.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.1 | 0.3 | GO:0002879 | positive regulation of acute inflammatory response to non-antigenic stimulus(GO:0002879) |
| 0.1 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.1 | 1.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.2 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.1 | 0.8 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 | 0.9 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 3.6 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.5 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.3 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 0.0 | 0.8 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.3 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.9 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.0 | 0.4 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.0 | 0.8 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.6 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.4 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0061143 | alveolar primary septum development(GO:0061143) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.6 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.5 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 4.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 2.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.9 | PID E2F PATHWAY | E2F transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 2.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 1.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.9 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.4 | 2.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.4 | 0.8 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.3 | 1.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.3 | 1.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.3 | 1.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.2 | 0.7 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 1.4 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 0.5 | GO:0036317 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.3 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 3.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.6 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.1 | 0.5 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0015361 | low-affinity sodium:dicarboxylate symporter activity(GO:0015361) |
| 0.1 | 0.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.8 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.5 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.6 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 2.4 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 2.3 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.6 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.2 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.6 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.6 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.9 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.5 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.0 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 1.3 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.6 | GO:0070288 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.1 | 0.5 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.5 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 3.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 1.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.9 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.1 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 1.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.5 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 2.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.6 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.9 | GO:1904724 | tertiary granule lumen(GO:1904724) |