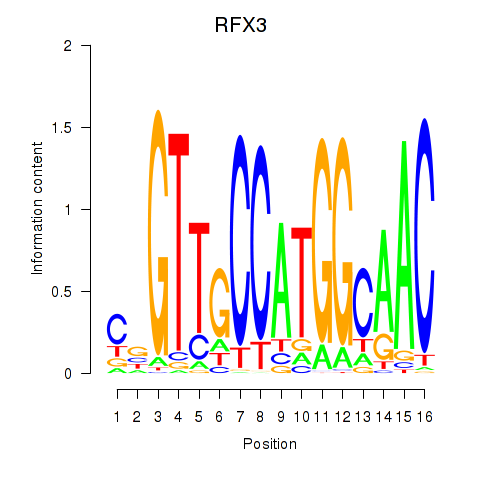
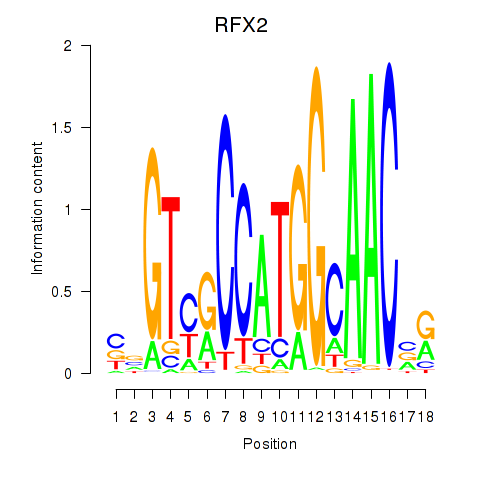
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for RFX3_RFX2
Z-value: 1.59
Transcription factors associated with RFX3_RFX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX3
|
ENSG00000080298.11 | RFX3 |
|
RFX2
|
ENSG00000087903.8 | RFX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX2 | hg19_v2_chr19_-_6110474_6110551 | 0.17 | 5.3e-01 | Click! |
| RFX3 | hg19_v2_chr9_-_3525968_3526016 | -0.05 | 8.5e-01 | Click! |
Activity profile of RFX3_RFX2 motif
Sorted Z-values of RFX3_RFX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX3_RFX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_133938820 | 6.76 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr11_+_133938955 | 6.17 |
ENST00000534549.1 ENST00000441717.3 |
JAM3 |
junctional adhesion molecule 3 |
| chr2_-_175870085 | 4.46 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr2_-_175869936 | 4.13 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr10_-_126694575 | 3.35 |
ENST00000334808.6 |
CTBP2 |
C-terminal binding protein 2 |
| chrX_+_135251783 | 3.29 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr14_+_105941118 | 2.83 |
ENST00000550577.1 ENST00000538259.2 |
CRIP2 |
cysteine-rich protein 2 |
| chr15_+_43803143 | 2.72 |
ENST00000382031.1 |
MAP1A |
microtubule-associated protein 1A |
| chr17_-_64187973 | 2.62 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr17_-_64188177 | 2.57 |
ENST00000535342.2 |
CEP112 |
centrosomal protein 112kDa |
| chr3_-_149688896 | 2.55 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr14_-_74181106 | 2.55 |
ENST00000316836.3 |
PNMA1 |
paraneoplastic Ma antigen 1 |
| chrX_+_135252050 | 2.45 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr19_-_58609570 | 2.15 |
ENST00000600845.1 ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18 |
zinc finger and SCAN domain containing 18 |
| chrX_+_135251835 | 2.12 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr11_+_844406 | 2.08 |
ENST00000397404.1 |
TSPAN4 |
tetraspanin 4 |
| chr17_-_76899275 | 2.03 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr20_+_43160458 | 1.89 |
ENST00000372889.1 ENST00000372887.1 ENST00000372882.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr20_+_43160409 | 1.87 |
ENST00000372894.3 ENST00000372892.3 ENST00000372891.3 |
PKIG |
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr14_+_100259666 | 1.81 |
ENST00000262233.6 ENST00000334192.4 |
EML1 |
echinoderm microtubule associated protein like 1 |
| chr6_+_52285131 | 1.73 |
ENST00000433625.2 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr15_+_96873921 | 1.61 |
ENST00000394166.3 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr6_+_52285046 | 1.59 |
ENST00000371068.5 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr15_+_71184931 | 1.57 |
ENST00000560369.1 ENST00000260382.5 |
LRRC49 |
leucine rich repeat containing 49 |
| chr5_-_179780312 | 1.51 |
ENST00000253778.8 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
| chrX_+_103173457 | 1.49 |
ENST00000419165.1 |
TMSB15B |
thymosin beta 15B |
| chr2_+_170335924 | 1.48 |
ENST00000554017.1 ENST00000392663.2 ENST00000513963.1 |
BBS5 RP11-724O16.1 |
Bardet-Biedl syndrome 5 Bardet-Biedl syndrome 5 protein; Uncharacterized protein |
| chr16_+_57279248 | 1.44 |
ENST00000562023.1 ENST00000563234.1 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr15_+_71185148 | 1.41 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr7_-_35077653 | 1.38 |
ENST00000310974.4 |
DPY19L1 |
dpy-19-like 1 (C. elegans) |
| chr1_+_84630053 | 1.38 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr1_-_169337176 | 1.36 |
ENST00000472647.1 ENST00000367811.3 |
NME7 |
NME/NM23 family member 7 |
| chr1_+_84630645 | 1.35 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr8_+_59465728 | 1.34 |
ENST00000260130.4 ENST00000422546.2 ENST00000447182.2 ENST00000413219.2 ENST00000424270.2 ENST00000523483.1 ENST00000520168.1 |
SDCBP |
syndecan binding protein (syntenin) |
| chr9_-_130635741 | 1.33 |
ENST00000223836.10 |
AK1 |
adenylate kinase 1 |
| chr5_-_16742330 | 1.28 |
ENST00000505695.1 ENST00000427430.2 |
MYO10 |
myosin X |
| chr14_-_102771462 | 1.23 |
ENST00000522874.1 |
MOK |
MOK protein kinase |
| chr3_-_107941230 | 1.22 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr22_+_39052632 | 1.22 |
ENST00000411557.1 ENST00000396811.2 ENST00000216029.3 ENST00000416285.1 |
CBY1 |
chibby homolog 1 (Drosophila) |
| chr15_-_43802769 | 1.19 |
ENST00000263801.3 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr8_-_98290087 | 1.18 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr6_-_116575226 | 1.17 |
ENST00000420283.1 |
TSPYL4 |
TSPY-like 4 |
| chr14_-_102771516 | 1.15 |
ENST00000524214.1 ENST00000193029.6 ENST00000361847.2 |
MOK |
MOK protein kinase |
| chr6_-_43484718 | 1.14 |
ENST00000372422.2 |
YIPF3 |
Yip1 domain family, member 3 |
| chr22_-_43485381 | 1.13 |
ENST00000331018.7 ENST00000266254.7 ENST00000445824.1 |
TTLL1 |
tubulin tyrosine ligase-like family, member 1 |
| chr16_-_88923285 | 1.11 |
ENST00000542788.1 ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS |
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr16_+_57279004 | 1.10 |
ENST00000219204.3 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
| chr17_+_26662730 | 1.04 |
ENST00000226225.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr1_-_59249732 | 1.04 |
ENST00000371222.2 |
JUN |
jun proto-oncogene |
| chr2_+_232063260 | 1.01 |
ENST00000349938.4 |
ARMC9 |
armadillo repeat containing 9 |
| chr14_-_24664776 | 0.98 |
ENST00000530468.1 ENST00000528010.1 ENST00000396854.4 ENST00000524835.1 ENST00000261789.4 ENST00000525592.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr11_+_102980126 | 0.98 |
ENST00000375735.2 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr6_-_43484621 | 0.98 |
ENST00000506469.1 ENST00000503972.1 |
YIPF3 |
Yip1 domain family, member 3 |
| chr19_-_47354082 | 0.96 |
ENST00000593442.1 ENST00000263270.6 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
| chr1_-_114302086 | 0.96 |
ENST00000369604.1 ENST00000357783.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr5_+_148521381 | 0.94 |
ENST00000504238.1 |
ABLIM3 |
actin binding LIM protein family, member 3 |
| chr1_+_38022572 | 0.94 |
ENST00000541606.1 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr11_-_67276100 | 0.93 |
ENST00000301488.3 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
| chr3_-_49055991 | 0.93 |
ENST00000441576.2 ENST00000420952.2 ENST00000341949.4 ENST00000395462.4 |
DALRD3 |
DALR anticodon binding domain containing 3 |
| chr1_-_114301755 | 0.90 |
ENST00000393357.2 ENST00000369596.2 ENST00000446739.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr12_-_131323754 | 0.86 |
ENST00000261653.6 |
STX2 |
syntaxin 2 |
| chr5_+_121647764 | 0.85 |
ENST00000261368.8 ENST00000379533.2 ENST00000379536.2 ENST00000379538.3 |
SNCAIP |
synuclein, alpha interacting protein |
| chr5_+_121647924 | 0.84 |
ENST00000414317.2 |
SNCAIP |
synuclein, alpha interacting protein |
| chr3_-_58563094 | 0.84 |
ENST00000464064.1 |
FAM107A |
family with sequence similarity 107, member A |
| chr14_-_24664540 | 0.84 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr11_+_66278080 | 0.83 |
ENST00000318312.7 ENST00000526815.1 ENST00000537537.1 ENST00000525809.1 ENST00000455748.2 ENST00000393994.2 |
BBS1 |
Bardet-Biedl syndrome 1 |
| chr1_-_114301960 | 0.83 |
ENST00000369598.1 ENST00000369600.1 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr10_+_70748487 | 0.83 |
ENST00000361983.4 |
KIAA1279 |
KIAA1279 |
| chr3_+_158288999 | 0.83 |
ENST00000482628.1 ENST00000478894.2 ENST00000392822.3 ENST00000466246.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr11_+_102980251 | 0.83 |
ENST00000334267.7 ENST00000398093.3 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr11_-_6426635 | 0.83 |
ENST00000608645.1 ENST00000608394.1 ENST00000529519.1 |
APBB1 |
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr19_-_47354023 | 0.83 |
ENST00000601649.1 ENST00000599990.1 ENST00000352203.4 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
| chr5_+_121647877 | 0.82 |
ENST00000514497.2 ENST00000261367.7 |
SNCAIP |
synuclein, alpha interacting protein |
| chr15_-_55700457 | 0.82 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chr14_-_23058063 | 0.81 |
ENST00000538631.1 ENST00000543337.1 ENST00000250498.4 |
DAD1 |
defender against cell death 1 |
| chr8_+_120428546 | 0.80 |
ENST00000259526.3 |
NOV |
nephroblastoma overexpressed |
| chr5_+_92919043 | 0.80 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr11_-_64546202 | 0.77 |
ENST00000377390.3 ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1 |
splicing factor 1 |
| chr4_-_186733363 | 0.76 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr17_+_26662597 | 0.74 |
ENST00000544907.2 |
TNFAIP1 |
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr19_+_3572758 | 0.73 |
ENST00000416526.1 |
HMG20B |
high mobility group 20B |
| chr15_-_55700216 | 0.72 |
ENST00000569205.1 |
CCPG1 |
cell cycle progression 1 |
| chr1_-_48937838 | 0.72 |
ENST00000371847.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr1_-_48937821 | 0.71 |
ENST00000396199.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr14_+_74486043 | 0.71 |
ENST00000464394.1 ENST00000394009.3 |
CCDC176 |
coiled-coil domain containing 176 |
| chr1_-_109584768 | 0.71 |
ENST00000357672.3 |
WDR47 |
WD repeat domain 47 |
| chr1_-_109584716 | 0.71 |
ENST00000531337.1 ENST00000529074.1 ENST00000369965.4 |
WDR47 |
WD repeat domain 47 |
| chr13_+_37005967 | 0.69 |
ENST00000440264.1 ENST00000449823.1 |
CCNA1 |
cyclin A1 |
| chr19_-_46974741 | 0.68 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr10_-_17659234 | 0.68 |
ENST00000466335.1 |
PTPLA |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member A |
| chr14_+_74416989 | 0.68 |
ENST00000334571.2 ENST00000554920.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr19_-_46974664 | 0.67 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr12_-_131323719 | 0.67 |
ENST00000392373.2 |
STX2 |
syntaxin 2 |
| chr12_+_9067123 | 0.66 |
ENST00000543824.1 |
PHC1 |
polyhomeotic homolog 1 (Drosophila) |
| chr19_-_47353547 | 0.66 |
ENST00000601498.1 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
| chr11_+_17298297 | 0.66 |
ENST00000529010.1 |
NUCB2 |
nucleobindin 2 |
| chr2_-_44588893 | 0.66 |
ENST00000409272.1 ENST00000410081.1 ENST00000541738.1 |
PREPL |
prolyl endopeptidase-like |
| chr1_-_183604794 | 0.65 |
ENST00000367534.1 ENST00000359856.6 ENST00000294742.6 |
ARPC5 |
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr4_-_156298028 | 0.64 |
ENST00000433024.1 ENST00000379248.2 |
MAP9 |
microtubule-associated protein 9 |
| chr19_+_9945962 | 0.64 |
ENST00000587625.1 ENST00000247970.4 ENST00000588695.1 |
PIN1 |
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
| chr7_+_100273736 | 0.63 |
ENST00000412215.1 ENST00000393924.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr6_+_31939608 | 0.63 |
ENST00000375331.2 ENST00000375333.2 |
STK19 |
serine/threonine kinase 19 |
| chr5_+_72112470 | 0.63 |
ENST00000447967.2 ENST00000523768.1 |
TNPO1 |
transportin 1 |
| chr14_-_75643296 | 0.62 |
ENST00000303575.4 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr1_-_109584608 | 0.62 |
ENST00000400794.3 ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47 |
WD repeat domain 47 |
| chr9_+_132815985 | 0.61 |
ENST00000372410.3 |
GPR107 |
G protein-coupled receptor 107 |
| chr15_-_55700522 | 0.60 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr1_+_169337172 | 0.60 |
ENST00000367807.3 ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1 |
basic leucine zipper nuclear factor 1 |
| chr3_-_49761337 | 0.60 |
ENST00000535833.1 ENST00000308388.6 ENST00000480687.1 ENST00000308375.6 |
AMIGO3 GMPPB |
adhesion molecule with Ig-like domain 3 GDP-mannose pyrophosphorylase B |
| chr17_-_26662440 | 0.59 |
ENST00000578122.1 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr12_+_9067327 | 0.59 |
ENST00000433083.2 ENST00000544916.1 ENST00000544539.1 ENST00000539063.1 |
PHC1 |
polyhomeotic homolog 1 (Drosophila) |
| chr19_+_3572925 | 0.59 |
ENST00000333651.6 ENST00000417382.1 ENST00000453933.1 ENST00000262949.7 |
HMG20B |
high mobility group 20B |
| chr19_-_15560730 | 0.58 |
ENST00000389282.4 ENST00000263381.7 |
WIZ |
widely interspaced zinc finger motifs |
| chr4_-_156297949 | 0.57 |
ENST00000515654.1 |
MAP9 |
microtubule-associated protein 9 |
| chr2_+_183580954 | 0.57 |
ENST00000264065.7 |
DNAJC10 |
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr11_+_17298255 | 0.57 |
ENST00000531172.1 ENST00000533738.2 ENST00000323688.6 |
NUCB2 |
nucleobindin 2 |
| chr1_+_38022513 | 0.56 |
ENST00000296218.7 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
| chr16_+_19729586 | 0.56 |
ENST00000564186.1 ENST00000541926.1 ENST00000433597.2 |
IQCK |
IQ motif containing K |
| chr6_+_30035307 | 0.55 |
ENST00000376765.2 ENST00000376763.1 |
PPP1R11 |
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr2_-_44588624 | 0.54 |
ENST00000438314.1 ENST00000409936.1 |
PREPL |
prolyl endopeptidase-like |
| chr6_+_30034865 | 0.53 |
ENST00000376772.3 |
PPP1R11 |
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chrX_+_129473916 | 0.52 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr1_-_48937682 | 0.52 |
ENST00000371843.3 |
SPATA6 |
spermatogenesis associated 6 |
| chr1_+_183605200 | 0.51 |
ENST00000304685.4 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr4_+_87515454 | 0.50 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr17_+_2240775 | 0.50 |
ENST00000268989.3 ENST00000426855.2 |
SGSM2 |
small G protein signaling modulator 2 |
| chr11_+_111807863 | 0.50 |
ENST00000440460.2 |
DIXDC1 |
DIX domain containing 1 |
| chr12_+_4671352 | 0.50 |
ENST00000542744.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_+_158288960 | 0.50 |
ENST00000484955.1 ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr21_-_35014027 | 0.50 |
ENST00000399442.1 ENST00000413017.2 ENST00000445393.1 ENST00000417979.1 ENST00000426935.1 ENST00000381540.3 ENST00000361534.2 ENST00000381554.3 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chr1_+_203444887 | 0.50 |
ENST00000343110.2 |
PRELP |
proline/arginine-rich end leucine-rich repeat protein |
| chr7_-_44229022 | 0.50 |
ENST00000403799.3 |
GCK |
glucokinase (hexokinase 4) |
| chr6_+_30034966 | 0.50 |
ENST00000376769.2 |
PPP1R11 |
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr10_-_17243579 | 0.50 |
ENST00000525762.1 ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
| chr5_+_72112418 | 0.49 |
ENST00000454282.1 |
TNPO1 |
transportin 1 |
| chr12_+_120933859 | 0.49 |
ENST00000242577.6 ENST00000548214.1 ENST00000392508.2 |
DYNLL1 |
dynein, light chain, LC8-type 1 |
| chr13_+_38923959 | 0.49 |
ENST00000379649.1 ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1 |
ubiquitin-fold modifier 1 |
| chr14_+_102829300 | 0.49 |
ENST00000359520.7 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
| chr17_+_41322483 | 0.48 |
ENST00000341165.6 ENST00000586650.1 ENST00000422280.1 |
NBR1 |
neighbor of BRCA1 gene 1 |
| chr2_-_44588679 | 0.48 |
ENST00000409411.1 |
PREPL |
prolyl endopeptidase-like |
| chr8_+_22462532 | 0.48 |
ENST00000389279.3 |
CCAR2 |
cell cycle and apoptosis regulator 2 |
| chr14_+_75469606 | 0.47 |
ENST00000266126.5 |
EIF2B2 |
eukaryotic translation initiation factor 2B, subunit 2 beta, 39kDa |
| chr17_-_56084578 | 0.47 |
ENST00000582730.2 ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1 |
serine/arginine-rich splicing factor 1 |
| chr8_-_101964231 | 0.46 |
ENST00000521309.1 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr16_+_57662419 | 0.45 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr22_-_36357671 | 0.45 |
ENST00000408983.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr17_-_26662464 | 0.45 |
ENST00000579419.1 ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr4_-_156298087 | 0.44 |
ENST00000311277.4 |
MAP9 |
microtubule-associated protein 9 |
| chr19_-_2783363 | 0.44 |
ENST00000221566.2 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr1_-_159893507 | 0.44 |
ENST00000368096.1 |
TAGLN2 |
transgelin 2 |
| chrX_+_129473859 | 0.44 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr7_+_99699280 | 0.43 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr3_-_52739762 | 0.43 |
ENST00000487642.1 ENST00000464705.1 ENST00000491606.1 ENST00000489119.1 ENST00000478968.2 |
GLT8D1 |
glycosyltransferase 8 domain containing 1 |
| chr2_-_44588694 | 0.43 |
ENST00000409957.1 |
PREPL |
prolyl endopeptidase-like |
| chr9_-_110251836 | 0.43 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr3_+_14166440 | 0.42 |
ENST00000306077.4 |
TMEM43 |
transmembrane protein 43 |
| chr12_+_120933904 | 0.42 |
ENST00000550178.1 ENST00000550845.1 ENST00000549989.1 ENST00000552870.1 |
DYNLL1 |
dynein, light chain, LC8-type 1 |
| chr19_+_13842559 | 0.42 |
ENST00000586600.1 |
CCDC130 |
coiled-coil domain containing 130 |
| chr3_+_49027308 | 0.42 |
ENST00000383729.4 ENST00000343546.4 |
P4HTM |
prolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) |
| chr17_-_61523535 | 0.42 |
ENST00000584031.1 ENST00000392976.1 |
CYB561 |
cytochrome b561 |
| chr13_+_37006421 | 0.41 |
ENST00000255465.4 |
CCNA1 |
cyclin A1 |
| chr16_+_3550924 | 0.40 |
ENST00000576634.1 ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1 |
clusterin associated protein 1 |
| chr1_-_44820880 | 0.40 |
ENST00000372257.2 ENST00000457571.1 ENST00000452396.1 |
ERI3 |
ERI1 exoribonuclease family member 3 |
| chr17_+_66508154 | 0.40 |
ENST00000358598.2 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chrX_-_102941596 | 0.39 |
ENST00000441076.2 ENST00000422355.1 ENST00000442614.1 ENST00000422154.2 ENST00000451301.1 |
MORF4L2 |
mortality factor 4 like 2 |
| chr3_-_121468513 | 0.39 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr20_-_20033052 | 0.39 |
ENST00000536226.1 |
CRNKL1 |
crooked neck pre-mRNA splicing factor 1 |
| chr6_-_32145861 | 0.39 |
ENST00000336984.6 |
AGPAT1 |
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr15_+_72978539 | 0.38 |
ENST00000539603.1 ENST00000569338.1 |
BBS4 |
Bardet-Biedl syndrome 4 |
| chr1_+_203595903 | 0.38 |
ENST00000367218.3 ENST00000367219.3 ENST00000391954.2 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr7_+_99699179 | 0.38 |
ENST00000438383.1 ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr6_-_135818844 | 0.38 |
ENST00000524469.1 ENST00000367800.4 ENST00000327035.6 ENST00000457866.2 ENST00000265602.6 |
AHI1 |
Abelson helper integration site 1 |
| chr8_+_22462145 | 0.38 |
ENST00000308511.4 ENST00000523801.1 ENST00000521301.1 |
CCAR2 |
cell cycle and apoptosis regulator 2 |
| chr14_+_57046530 | 0.38 |
ENST00000536419.1 ENST00000538838.1 |
TMEM260 |
transmembrane protein 260 |
| chr10_-_119806085 | 0.38 |
ENST00000355624.3 |
RAB11FIP2 |
RAB11 family interacting protein 2 (class I) |
| chr13_+_37006398 | 0.37 |
ENST00000418263.1 |
CCNA1 |
cyclin A1 |
| chr19_+_36605850 | 0.37 |
ENST00000221855.3 |
TBCB |
tubulin folding cofactor B |
| chr20_-_44540686 | 0.37 |
ENST00000477313.1 ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP |
phospholipid transfer protein |
| chr7_-_45128472 | 0.37 |
ENST00000490531.2 |
NACAD |
NAC alpha domain containing |
| chr11_-_64545941 | 0.37 |
ENST00000377387.1 |
SF1 |
splicing factor 1 |
| chr11_+_61447845 | 0.37 |
ENST00000257215.5 |
DAGLA |
diacylglycerol lipase, alpha |
| chr1_-_217804377 | 0.36 |
ENST00000366935.3 ENST00000366934.3 |
GPATCH2 |
G patch domain containing 2 |
| chr15_+_72978521 | 0.36 |
ENST00000542334.1 ENST00000268057.4 |
BBS4 |
Bardet-Biedl syndrome 4 |
| chr7_+_106685079 | 0.35 |
ENST00000265717.4 |
PRKAR2B |
protein kinase, cAMP-dependent, regulatory, type II, beta |
| chr3_-_52740012 | 0.35 |
ENST00000407584.3 ENST00000266014.5 |
GLT8D1 |
glycosyltransferase 8 domain containing 1 |
| chr2_-_47403642 | 0.35 |
ENST00000456319.1 ENST00000409563.1 ENST00000272298.7 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
| chr2_+_237994519 | 0.35 |
ENST00000392008.2 ENST00000409334.1 ENST00000409629.1 |
COPS8 |
COP9 signalosome subunit 8 |
| chr12_-_49351228 | 0.35 |
ENST00000541959.1 ENST00000447318.2 |
ARF3 |
ADP-ribosylation factor 3 |
| chr15_-_43785303 | 0.35 |
ENST00000382039.3 ENST00000450115.2 ENST00000382044.4 |
TP53BP1 |
tumor protein p53 binding protein 1 |
| chr7_+_100797726 | 0.34 |
ENST00000429457.1 |
AP1S1 |
adaptor-related protein complex 1, sigma 1 subunit |
| chr12_+_53693466 | 0.33 |
ENST00000267103.5 ENST00000548632.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr7_+_150929550 | 0.33 |
ENST00000482173.1 ENST00000495645.1 ENST00000035307.2 |
CHPF2 |
chondroitin polymerizing factor 2 |
| chr17_-_4167142 | 0.33 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr17_+_66508537 | 0.33 |
ENST00000392711.1 ENST00000585427.1 ENST00000589228.1 ENST00000536854.2 ENST00000588702.1 ENST00000589309.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr1_-_114301503 | 0.33 |
ENST00000447664.2 |
PHTF1 |
putative homeodomain transcription factor 1 |
| chr10_-_120101804 | 0.32 |
ENST00000369183.4 ENST00000369172.4 |
FAM204A |
family with sequence similarity 204, member A |
| chr1_+_203595689 | 0.32 |
ENST00000357681.5 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr19_-_50370509 | 0.32 |
ENST00000596014.1 |
PNKP |
polynucleotide kinase 3'-phosphatase |
| chr3_-_121468602 | 0.32 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr1_+_203651937 | 0.32 |
ENST00000341360.2 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
Gene Ontology Analysis
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 12.9 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.5 | 1.9 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.4 | 5.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 1.0 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 3.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.3 | 1.5 | GO:0097123 | cyclin A1-CDK2 complex(GO:0097123) |
| 0.3 | 0.9 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.3 | 3.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.2 | 3.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.2 | 4.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.5 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.8 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 1.0 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 2.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.1 | 2.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.6 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.8 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 2.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.4 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 1.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 3.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 0.5 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 1.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 0.4 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.1 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 3.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 1.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 2.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.7 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 2.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 14.7 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.9 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.5 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 3.6 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.4 | GO:0043202 | lysosomal lumen(GO:0043202) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 4.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.1 | 2.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 12.8 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.1 | 1.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.5 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 1.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 8.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.7 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 1.0 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.6 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 2.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.5 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.3 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.6 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 13.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.6 | 3.1 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.5 | 1.4 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.4 | 2.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.4 | 1.6 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.4 | 3.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.3 | 1.0 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.3 | 0.9 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.3 | 4.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.3 | 0.8 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.3 | 1.3 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.3 | 0.8 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.3 | 1.0 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.3 | 8.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.2 | 0.7 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 1.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.2 | 2.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.2 | 2.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 1.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 0.9 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.2 | 0.4 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.2 | 1.7 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.2 | 0.9 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.2 | 0.6 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 5.2 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.6 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.4 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.4 | GO:0071409 | negative regulation of muscle hyperplasia(GO:0014740) cellular response to cycloheximide(GO:0071409) |
| 0.1 | 7.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 2.4 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.1 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.1 | 0.6 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.1 | 1.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 1.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.4 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.1 | 2.5 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.1 | 1.6 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.1 | 2.8 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.1 | 2.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 1.5 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 1.8 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.3 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 3.8 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.1 | 0.9 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.5 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.2 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 0.5 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 1.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 2.1 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.1 | 0.7 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.2 | GO:0070213 | negative regulation of sister chromatid cohesion(GO:0045875) protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 1.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.3 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.6 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 3.0 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.1 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 2.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0019605 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.7 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:1902903 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) regulation of fibril organization(GO:1902903) negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 1.7 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.8 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 1.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.5 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.8 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:1903305 | regulation of calcium ion-dependent exocytosis(GO:0017158) regulation of regulated secretory pathway(GO:1903305) |
| 0.0 | 0.5 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 1.3 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.2 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 1.6 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.4 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 1.2 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.3 | GO:0071028 | nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.6 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.8 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.3 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.4 | GO:0003091 | renal water homeostasis(GO:0003091) |
| 0.0 | 0.0 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.0 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 1.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0050774 | manganese ion transport(GO:0006828) negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.5 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 12.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.1 | 7.0 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.7 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.5 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 4.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 2.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 1.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.4 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.4 | 1.3 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.3 | 1.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.3 | 0.9 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.3 | 1.3 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 1.5 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.2 | 4.8 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 1.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 1.0 | GO:0002046 | opsin binding(GO:0002046) |
| 0.2 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.6 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.2 | 2.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.5 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.2 | 0.7 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.2 | 1.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 8.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 0.6 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.1 | 0.6 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.1 | 0.6 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.8 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.1 | 2.7 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 2.5 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.3 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.8 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 1.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.6 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 2.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 0.3 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.1 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 14.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.5 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.2 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.7 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.7 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 0.5 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.3 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 1.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 0.6 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 1.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 2.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 6.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 1.6 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.7 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.5 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 1.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.6 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |