Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for RFX7_RFX4_RFX1
Z-value: 1.10
Transcription factors associated with RFX7_RFX4_RFX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
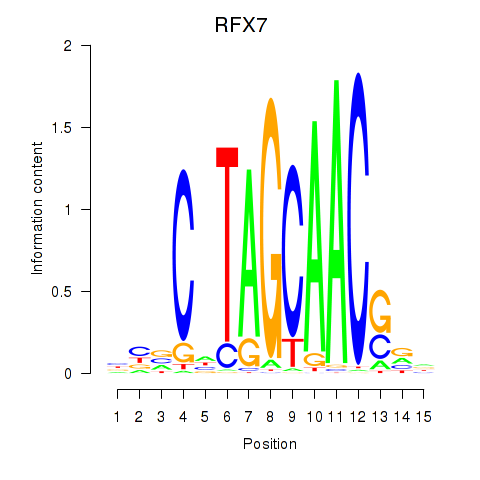
RFX7
|
ENSG00000181827.10 | RFX7 |
|
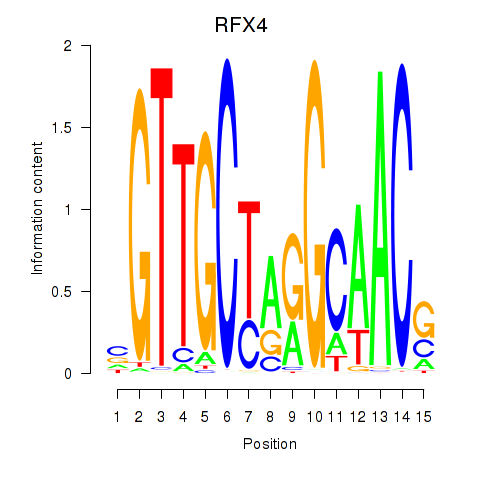
RFX4
|
ENSG00000111783.8 | RFX4 |
|
RFX1
|
ENSG00000132005.4 | RFX1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RFX7 | hg19_v2_chr15_-_56535464_56535521 | -0.32 | 2.3e-01 | Click! |
| RFX1 | hg19_v2_chr19_-_14117074_14117141 | 0.22 | 4.2e-01 | Click! |
Activity profile of RFX7_RFX4_RFX1 motif
Sorted Z-values of RFX7_RFX4_RFX1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RFX7_RFX4_RFX1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_15397732 | 4.26 |
ENST00000382020.4 ENST00000506802.1 ENST00000509380.1 ENST00000503731.1 |
TUSC3 |
tumor suppressor candidate 3 |
| chr4_-_156298028 | 2.43 |
ENST00000433024.1 ENST00000379248.2 |
MAP9 |
microtubule-associated protein 9 |
| chrX_+_135251783 | 1.94 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr4_-_156297949 | 1.84 |
ENST00000515654.1 |
MAP9 |
microtubule-associated protein 9 |
| chr4_-_156298087 | 1.81 |
ENST00000311277.4 |
MAP9 |
microtubule-associated protein 9 |
| chr14_-_102771516 | 1.68 |
ENST00000524214.1 ENST00000193029.6 ENST00000361847.2 |
MOK |
MOK protein kinase |
| chr14_-_102771462 | 1.63 |
ENST00000522874.1 |
MOK |
MOK protein kinase |
| chrX_+_135252050 | 1.60 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr19_-_45953983 | 1.58 |
ENST00000592083.1 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chrX_+_135251835 | 1.40 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr12_+_13349650 | 1.37 |
ENST00000256951.5 ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1 |
epithelial membrane protein 1 |
| chr4_+_26585686 | 1.36 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr4_+_26585538 | 1.30 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr20_+_17550691 | 1.18 |
ENST00000474024.1 |
DSTN |
destrin (actin depolymerizing factor) |
| chr2_-_190044480 | 1.16 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr1_+_150480551 | 1.14 |
ENST00000369049.4 ENST00000369047.4 |
ECM1 |
extracellular matrix protein 1 |
| chr11_-_62521614 | 1.11 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chrX_-_114252193 | 1.08 |
ENST00000243213.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chrX_+_47444613 | 1.07 |
ENST00000445623.1 |
TIMP1 |
TIMP metallopeptidase inhibitor 1 |
| chr11_+_69455855 | 1.06 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr1_+_150480576 | 1.06 |
ENST00000346569.6 |
ECM1 |
extracellular matrix protein 1 |
| chr20_+_17550489 | 1.03 |
ENST00000246069.7 |
DSTN |
destrin (actin depolymerizing factor) |
| chr10_-_95241951 | 1.02 |
ENST00000358334.5 ENST00000359263.4 ENST00000371488.3 |
MYOF |
myoferlin |
| chr10_-_95242044 | 0.99 |
ENST00000371501.4 ENST00000371502.4 ENST00000371489.1 |
MYOF |
myoferlin |
| chr12_-_47473707 | 0.96 |
ENST00000429635.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr11_+_113185533 | 0.96 |
ENST00000393020.1 |
TTC12 |
tetratricopeptide repeat domain 12 |
| chr1_-_85156090 | 0.92 |
ENST00000605755.1 ENST00000437941.2 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr16_-_84538218 | 0.92 |
ENST00000562447.1 ENST00000565765.1 ENST00000535580.1 ENST00000343629.6 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
| chr9_-_130635741 | 0.89 |
ENST00000223836.10 |
AK1 |
adenylate kinase 1 |
| chr1_-_85156216 | 0.88 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr11_-_108422926 | 0.88 |
ENST00000428840.1 ENST00000526312.1 |
EXPH5 |
exophilin 5 |
| chr19_-_58609570 | 0.87 |
ENST00000600845.1 ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18 |
zinc finger and SCAN domain containing 18 |
| chr11_+_113185251 | 0.86 |
ENST00000529221.1 |
TTC12 |
tetratricopeptide repeat domain 12 |
| chr8_+_59323823 | 0.80 |
ENST00000399598.2 |
UBXN2B |
UBX domain protein 2B |
| chr6_+_116601265 | 0.78 |
ENST00000452085.3 |
DSE |
dermatan sulfate epimerase |
| chr7_+_55177416 | 0.71 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr15_+_71184931 | 0.70 |
ENST00000560369.1 ENST00000260382.5 |
LRRC49 |
leucine rich repeat containing 49 |
| chr9_-_110251836 | 0.67 |
ENST00000374672.4 |
KLF4 |
Kruppel-like factor 4 (gut) |
| chr2_-_175869936 | 0.66 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr2_-_175870085 | 0.66 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr3_-_123339418 | 0.64 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr15_+_71185148 | 0.63 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr3_-_123339343 | 0.62 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chrX_-_13835147 | 0.55 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr11_+_113185292 | 0.53 |
ENST00000429951.1 ENST00000442859.1 ENST00000531164.1 ENST00000529850.1 ENST00000314756.3 ENST00000525965.1 |
TTC12 |
tetratricopeptide repeat domain 12 |
| chr11_-_119991589 | 0.51 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr2_+_170335924 | 0.51 |
ENST00000554017.1 ENST00000392663.2 ENST00000513963.1 |
BBS5 RP11-724O16.1 |
Bardet-Biedl syndrome 5 Bardet-Biedl syndrome 5 protein; Uncharacterized protein |
| chr1_+_203595903 | 0.50 |
ENST00000367218.3 ENST00000367219.3 ENST00000391954.2 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr5_+_110074685 | 0.50 |
ENST00000355943.3 ENST00000447245.2 |
SLC25A46 |
solute carrier family 25, member 46 |
| chr11_+_133938820 | 0.49 |
ENST00000299106.4 ENST00000529443.2 |
JAM3 |
junctional adhesion molecule 3 |
| chr1_+_144220127 | 0.48 |
ENST00000369373.5 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr12_-_49582978 | 0.48 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr11_+_133938955 | 0.46 |
ENST00000534549.1 ENST00000441717.3 |
JAM3 |
junctional adhesion molecule 3 |
| chr14_-_67826486 | 0.46 |
ENST00000555431.1 ENST00000554236.1 ENST00000555474.1 |
ATP6V1D |
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr11_-_66103867 | 0.46 |
ENST00000424433.2 |
RIN1 |
Ras and Rab interactor 1 |
| chr1_+_203595689 | 0.45 |
ENST00000357681.5 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
| chr20_+_48807351 | 0.44 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr3_+_135741576 | 0.44 |
ENST00000334546.2 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
| chr11_-_66112555 | 0.44 |
ENST00000425825.2 ENST00000359957.3 |
BRMS1 |
breast cancer metastasis suppressor 1 |
| chr2_+_242088963 | 0.44 |
ENST00000438799.1 ENST00000402734.1 ENST00000423280.1 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr16_+_56691606 | 0.44 |
ENST00000334350.6 |
MT1F |
metallothionein 1F |
| chr5_+_271752 | 0.42 |
ENST00000505221.1 ENST00000509581.1 ENST00000507528.1 |
PDCD6 |
programmed cell death 6 |
| chr1_-_43638168 | 0.42 |
ENST00000431635.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr1_+_206808918 | 0.40 |
ENST00000367108.3 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr19_-_39390440 | 0.40 |
ENST00000249396.7 ENST00000414941.1 ENST00000392081.2 |
SIRT2 |
sirtuin 2 |
| chr19_+_751122 | 0.39 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr1_+_206809113 | 0.39 |
ENST00000441486.1 ENST00000367106.1 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr1_+_206808868 | 0.38 |
ENST00000367109.2 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
| chr2_+_242089046 | 0.37 |
ENST00000407025.1 ENST00000272983.8 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr14_-_24664776 | 0.37 |
ENST00000530468.1 ENST00000528010.1 ENST00000396854.4 ENST00000524835.1 ENST00000261789.4 ENST00000525592.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr14_-_61116168 | 0.36 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr5_-_180237445 | 0.36 |
ENST00000393340.3 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr18_-_33702078 | 0.36 |
ENST00000586829.1 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr19_-_39390350 | 0.35 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr8_-_98290087 | 0.35 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr8_+_10530155 | 0.35 |
ENST00000521818.1 |
C8orf74 |
chromosome 8 open reading frame 74 |
| chr1_-_159893507 | 0.35 |
ENST00000368096.1 |
TAGLN2 |
transgelin 2 |
| chr9_+_19049372 | 0.34 |
ENST00000380527.1 |
RRAGA |
Ras-related GTP binding A |
| chr2_-_31361543 | 0.34 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr6_+_52285131 | 0.33 |
ENST00000433625.2 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr14_-_24664540 | 0.33 |
ENST00000530563.1 ENST00000528895.1 ENST00000528669.1 ENST00000532632.1 |
TM9SF1 |
transmembrane 9 superfamily member 1 |
| chr3_-_107941230 | 0.33 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr14_-_88459503 | 0.33 |
ENST00000393568.4 ENST00000261304.2 |
GALC |
galactosylceramidase |
| chr11_+_73882144 | 0.33 |
ENST00000328257.8 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr11_+_124543694 | 0.33 |
ENST00000227135.2 ENST00000532692.1 |
SPA17 |
sperm autoantigenic protein 17 |
| chr2_+_220110177 | 0.33 |
ENST00000409638.3 ENST00000396738.2 ENST00000409516.3 |
STK16 |
serine/threonine kinase 16 |
| chr16_+_3550924 | 0.33 |
ENST00000576634.1 ENST00000574369.1 ENST00000341633.5 ENST00000417763.2 ENST00000571025.1 |
CLUAP1 |
clusterin associated protein 1 |
| chr17_+_17876127 | 0.33 |
ENST00000582416.1 ENST00000313838.8 ENST00000411504.2 ENST00000581264.1 ENST00000399187.1 ENST00000479684.2 ENST00000584166.1 ENST00000585108.1 ENST00000399182.1 ENST00000579977.1 |
LRRC48 |
leucine rich repeat containing 48 |
| chr16_+_57673430 | 0.32 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr12_-_105629852 | 0.32 |
ENST00000551662.1 ENST00000553097.1 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr11_-_10562710 | 0.32 |
ENST00000528665.1 ENST00000265981.2 |
RNF141 |
ring finger protein 141 |
| chr11_+_73882311 | 0.32 |
ENST00000398427.4 ENST00000544401.1 |
PPME1 |
protein phosphatase methylesterase 1 |
| chr2_-_220110111 | 0.32 |
ENST00000428427.1 ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L |
galactosidase, beta 1-like |
| chr19_-_46974741 | 0.31 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr9_+_26956371 | 0.31 |
ENST00000380062.5 ENST00000518614.1 |
IFT74 |
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr19_-_46974664 | 0.31 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr15_+_49913201 | 0.31 |
ENST00000329873.5 ENST00000558653.1 ENST00000559164.1 ENST00000560632.1 ENST00000559405.1 ENST00000251250.6 |
DTWD1 |
DTW domain containing 1 |
| chr17_-_17875688 | 0.31 |
ENST00000379504.3 ENST00000318094.10 ENST00000540946.1 ENST00000542206.1 ENST00000395739.4 ENST00000581396.1 ENST00000535933.1 ENST00000579586.1 |
TOM1L2 |
target of myb1-like 2 (chicken) |
| chr1_+_211432775 | 0.30 |
ENST00000419091.2 |
RCOR3 |
REST corepressor 3 |
| chr2_-_9143786 | 0.29 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr14_-_23388338 | 0.29 |
ENST00000555209.1 ENST00000554256.1 ENST00000557403.1 ENST00000557549.1 ENST00000555676.1 ENST00000557571.1 ENST00000557464.1 ENST00000554618.1 ENST00000556862.1 ENST00000555722.1 ENST00000346528.5 ENST00000542016.2 ENST00000399922.2 ENST00000557227.1 ENST00000359890.3 |
RBM23 |
RNA binding motif protein 23 |
| chr14_+_105452094 | 0.29 |
ENST00000551606.1 ENST00000547315.1 |
C14orf79 |
chromosome 14 open reading frame 79 |
| chr4_+_668348 | 0.29 |
ENST00000511290.1 |
MYL5 |
myosin, light chain 5, regulatory |
| chr7_-_137028534 | 0.29 |
ENST00000348225.2 |
PTN |
pleiotrophin |
| chr15_-_85197501 | 0.29 |
ENST00000434634.2 |
WDR73 |
WD repeat domain 73 |
| chr6_-_43484718 | 0.29 |
ENST00000372422.2 |
YIPF3 |
Yip1 domain family, member 3 |
| chr12_-_105630016 | 0.29 |
ENST00000258530.3 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr15_+_67430339 | 0.29 |
ENST00000439724.3 |
SMAD3 |
SMAD family member 3 |
| chr6_+_52285046 | 0.29 |
ENST00000371068.5 |
EFHC1 |
EF-hand domain (C-terminal) containing 1 |
| chr15_+_49913175 | 0.29 |
ENST00000403028.3 |
DTWD1 |
DTW domain containing 1 |
| chr20_-_30311703 | 0.28 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr18_+_54318566 | 0.28 |
ENST00000589935.1 ENST00000357574.3 |
WDR7 |
WD repeat domain 7 |
| chr22_+_31477296 | 0.28 |
ENST00000426927.1 ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN |
smoothelin |
| chr19_+_52693259 | 0.28 |
ENST00000322088.6 ENST00000454220.2 ENST00000444322.2 ENST00000477989.1 |
PPP2R1A |
protein phosphatase 2, regulatory subunit A, alpha |
| chr13_+_21141270 | 0.28 |
ENST00000319980.6 ENST00000537103.1 ENST00000389373.3 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr17_-_64187973 | 0.27 |
ENST00000583358.1 ENST00000392769.2 |
CEP112 |
centrosomal protein 112kDa |
| chr2_-_110371412 | 0.27 |
ENST00000415095.1 ENST00000334001.6 ENST00000437928.1 ENST00000493445.1 ENST00000397714.2 ENST00000461295.1 |
SEPT10 |
septin 10 |
| chr3_-_123304017 | 0.27 |
ENST00000383657.5 |
PTPLB |
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b |
| chr2_-_110371777 | 0.27 |
ENST00000397712.2 |
SEPT10 |
septin 10 |
| chr17_-_80656528 | 0.27 |
ENST00000538809.2 ENST00000269347.6 ENST00000571995.1 |
RAB40B |
RAB40B, member RAS oncogene family |
| chr1_-_6420737 | 0.27 |
ENST00000541130.1 ENST00000377845.3 |
ACOT7 |
acyl-CoA thioesterase 7 |
| chrX_+_154114635 | 0.27 |
ENST00000369446.2 |
F8A1 |
coagulation factor VIII-associated 1 |
| chr11_-_64546202 | 0.27 |
ENST00000377390.3 ENST00000227503.9 ENST00000377394.3 ENST00000422298.2 ENST00000334944.5 |
SF1 |
splicing factor 1 |
| chr11_-_118436707 | 0.26 |
ENST00000264020.2 ENST00000264021.3 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr11_-_118436606 | 0.26 |
ENST00000530872.1 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr2_-_220110187 | 0.26 |
ENST00000295759.7 ENST00000392089.2 |
GLB1L |
galactosidase, beta 1-like |
| chr11_-_3078616 | 0.26 |
ENST00000401769.3 ENST00000278224.9 ENST00000397114.3 ENST00000380525.4 |
CARS |
cysteinyl-tRNA synthetase |
| chrX_-_154688276 | 0.26 |
ENST00000369445.2 |
F8A3 |
coagulation factor VIII-associated 3 |
| chr6_-_116601044 | 0.26 |
ENST00000368608.3 |
TSPYL1 |
TSPY-like 1 |
| chr11_+_65779283 | 0.26 |
ENST00000312134.2 |
CST6 |
cystatin E/M |
| chr13_+_21141208 | 0.25 |
ENST00000351808.5 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr18_+_54318616 | 0.25 |
ENST00000254442.3 |
WDR7 |
WD repeat domain 7 |
| chr3_+_111718036 | 0.25 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr17_-_64188177 | 0.25 |
ENST00000535342.2 |
CEP112 |
centrosomal protein 112kDa |
| chr1_+_1447517 | 0.25 |
ENST00000378756.3 ENST00000378755.5 |
ATAD3A |
ATPase family, AAA domain containing 3A |
| chr1_+_181003067 | 0.25 |
ENST00000434571.2 ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1 |
major histocompatibility complex, class I-related |
| chr12_+_111051902 | 0.25 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr16_-_88923285 | 0.25 |
ENST00000542788.1 ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS |
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr11_+_63753883 | 0.25 |
ENST00000538426.1 ENST00000543004.1 |
OTUB1 |
OTU domain, ubiquitin aldehyde binding 1 |
| chr5_-_43313574 | 0.24 |
ENST00000325110.6 ENST00000433297.2 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr14_+_74416989 | 0.24 |
ENST00000334571.2 ENST00000554920.1 |
COQ6 |
coenzyme Q6 monooxygenase |
| chr12_-_125401885 | 0.24 |
ENST00000542416.1 |
UBC |
ubiquitin C |
| chr2_-_110371720 | 0.24 |
ENST00000356688.4 |
SEPT10 |
septin 10 |
| chrX_+_154611749 | 0.24 |
ENST00000369505.3 |
F8A2 |
coagulation factor VIII-associated 2 |
| chr12_+_111051832 | 0.24 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr7_+_155090271 | 0.23 |
ENST00000476756.1 |
INSIG1 |
insulin induced gene 1 |
| chr6_-_167275991 | 0.23 |
ENST00000510118.1 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
| chr6_-_43484621 | 0.23 |
ENST00000506469.1 ENST00000503972.1 |
YIPF3 |
Yip1 domain family, member 3 |
| chr16_+_16429787 | 0.23 |
ENST00000331436.4 ENST00000541593.1 |
AC138969.4 |
Protein PKD1P1 |
| chr22_+_23487513 | 0.23 |
ENST00000263116.2 ENST00000341989.4 |
RAB36 |
RAB36, member RAS oncogene family |
| chr2_+_84743562 | 0.23 |
ENST00000389394.3 ENST00000398278.2 |
DNAH6 |
dynein, axonemal, heavy chain 6 |
| chr1_-_21044489 | 0.23 |
ENST00000247986.2 |
KIF17 |
kinesin family member 17 |
| chr19_-_40324255 | 0.22 |
ENST00000593685.1 ENST00000600611.1 |
DYRK1B |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr17_-_76899275 | 0.22 |
ENST00000322630.2 ENST00000586713.1 |
DDC8 |
Protein DDC8 homolog |
| chr7_+_158649242 | 0.22 |
ENST00000407559.3 |
WDR60 |
WD repeat domain 60 |
| chr6_-_159065741 | 0.22 |
ENST00000367085.3 ENST00000367089.3 |
DYNLT1 |
dynein, light chain, Tctex-type 1 |
| chr3_+_111717600 | 0.22 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr2_+_214149113 | 0.21 |
ENST00000331683.5 ENST00000432529.2 ENST00000413312.1 ENST00000272898.7 ENST00000447990.1 |
SPAG16 |
sperm associated antigen 16 |
| chr15_+_42066632 | 0.21 |
ENST00000457542.2 ENST00000221214.6 ENST00000260357.7 ENST00000456763.2 |
MAPKBP1 |
mitogen-activated protein kinase binding protein 1 |
| chr1_-_217804377 | 0.21 |
ENST00000366935.3 ENST00000366934.3 |
GPATCH2 |
G patch domain containing 2 |
| chr2_+_238767517 | 0.21 |
ENST00000404910.2 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
| chr22_-_36357671 | 0.21 |
ENST00000408983.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr20_-_49575081 | 0.20 |
ENST00000371588.5 ENST00000371582.4 |
DPM1 |
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr1_+_207226574 | 0.20 |
ENST00000367080.3 ENST00000367079.2 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr19_+_12848299 | 0.20 |
ENST00000357332.3 |
ASNA1 |
arsA arsenite transporter, ATP-binding, homolog 1 (bacterial) |
| chr11_-_3078838 | 0.20 |
ENST00000397111.5 |
CARS |
cysteinyl-tRNA synthetase |
| chr20_-_49575058 | 0.20 |
ENST00000371584.4 ENST00000371583.5 ENST00000413082.1 |
DPM1 |
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr3_+_111717511 | 0.20 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr2_-_44588893 | 0.20 |
ENST00000409272.1 ENST00000410081.1 ENST00000541738.1 |
PREPL |
prolyl endopeptidase-like |
| chr19_-_10679697 | 0.20 |
ENST00000335766.2 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr3_-_121468513 | 0.20 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr5_-_143550159 | 0.20 |
ENST00000448443.2 ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5 |
Yip1 domain family, member 5 |
| chr19_-_51523412 | 0.20 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr3_-_37216055 | 0.20 |
ENST00000336686.4 |
LRRFIP2 |
leucine rich repeat (in FLII) interacting protein 2 |
| chr10_-_99161033 | 0.20 |
ENST00000315563.6 ENST00000370992.4 ENST00000414986.1 |
RRP12 |
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr5_+_271733 | 0.20 |
ENST00000264933.4 |
PDCD6 |
programmed cell death 6 |
| chr2_+_27851863 | 0.19 |
ENST00000264718.3 ENST00000610189.1 |
GPN1 |
GPN-loop GTPase 1 |
| chr21_-_44846999 | 0.19 |
ENST00000270162.6 |
SIK1 |
salt-inducible kinase 1 |
| chr14_+_61447927 | 0.19 |
ENST00000451406.1 |
SLC38A6 |
solute carrier family 38, member 6 |
| chr14_-_75643296 | 0.19 |
ENST00000303575.4 |
TMED10 |
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr22_+_38453207 | 0.19 |
ENST00000404072.3 ENST00000424694.1 |
PICK1 |
protein interacting with PRKCA 1 |
| chr17_-_56084578 | 0.18 |
ENST00000582730.2 ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1 |
serine/arginine-rich splicing factor 1 |
| chr16_+_15031300 | 0.18 |
ENST00000328085.6 |
NPIPA1 |
nuclear pore complex interacting protein family, member A1 |
| chr5_-_143550241 | 0.18 |
ENST00000522203.1 |
YIPF5 |
Yip1 domain family, member 5 |
| chr14_+_61447832 | 0.18 |
ENST00000354886.2 ENST00000267488.4 |
SLC38A6 |
solute carrier family 38, member 6 |
| chr11_+_112832133 | 0.18 |
ENST00000524665.1 |
NCAM1 |
neural cell adhesion molecule 1 |
| chr1_+_15736359 | 0.18 |
ENST00000375980.4 |
EFHD2 |
EF-hand domain family, member D2 |
| chr2_+_239229129 | 0.18 |
ENST00000391994.2 |
TRAF3IP1 |
TNF receptor-associated factor 3 interacting protein 1 |
| chr16_-_11363178 | 0.18 |
ENST00000312693.3 |
TNP2 |
transition protein 2 (during histone to protamine replacement) |
| chr5_-_54529415 | 0.18 |
ENST00000282572.4 |
CCNO |
cyclin O |
| chr16_+_14805546 | 0.18 |
ENST00000552140.1 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr5_+_68530668 | 0.18 |
ENST00000506563.1 |
CDK7 |
cyclin-dependent kinase 7 |
| chr11_-_507184 | 0.18 |
ENST00000533410.1 ENST00000354420.2 ENST00000397604.3 ENST00000531149.1 ENST00000356187.5 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr5_+_49963239 | 0.17 |
ENST00000505554.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr13_+_38923959 | 0.17 |
ENST00000379649.1 ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1 |
ubiquitin-fold modifier 1 |
| chr13_-_30881621 | 0.17 |
ENST00000380615.3 |
KATNAL1 |
katanin p60 subunit A-like 1 |
| chr1_-_45140227 | 0.17 |
ENST00000372237.3 |
TMEM53 |
transmembrane protein 53 |
| chr22_+_38453378 | 0.17 |
ENST00000437453.1 ENST00000356976.3 |
PICK1 |
protein interacting with PRKCA 1 |
| chr11_-_506739 | 0.16 |
ENST00000529306.1 ENST00000438658.2 ENST00000527485.1 ENST00000397615.2 ENST00000397614.1 |
RNH1 |
ribonuclease/angiogenin inhibitor 1 |
| chr1_-_183604794 | 0.16 |
ENST00000367534.1 ENST00000359856.6 ENST00000294742.6 |
ARPC5 |
actin related protein 2/3 complex, subunit 5, 16kDa |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.5 | 4.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 1.0 | GO:0033241 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.3 | 2.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.3 | 1.2 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.2 | 0.7 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.2 | 0.7 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.2 | 1.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.2 | 2.2 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.5 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.2 | 1.9 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.2 | 1.3 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.2 | 0.5 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.2 | 1.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 1.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.2 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.1 | 1.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.5 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.4 | GO:0030910 | olfactory placode formation(GO:0030910) myotome development(GO:0061055) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.2 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 1.9 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 0.3 | GO:1904395 | retinal rod cell differentiation(GO:0060221) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 5.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 2.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 1.4 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.4 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.2 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.2 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) |
| 0.1 | 0.1 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.3 | GO:1900533 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 0.3 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.1 | 0.6 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.1 | 0.4 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.1 | 0.5 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.2 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.1 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.1 | 0.3 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 1.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.4 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 1.0 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 1.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.2 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 1.3 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.9 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.4 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 2.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0032383 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.4 | GO:0019374 | galactosylceramide metabolic process(GO:0006681) galactolipid metabolic process(GO:0019374) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.9 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.0 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.5 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 1.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.7 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.2 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.2 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.8 | GO:0015949 | nucleobase-containing small molecule interconversion(GO:0015949) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) positive regulation of beta-amyloid clearance(GO:1900223) |
| 0.0 | 0.5 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.1 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0098838 | methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.1 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.6 | GO:0021795 | cerebral cortex cell migration(GO:0021795) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) |
| 0.0 | 0.1 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.4 | 2.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.3 | 1.0 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.3 | 0.8 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.3 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 1.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 0.5 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.1 | 0.7 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.6 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 4.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 0.3 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.6 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.8 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.3 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.3 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0008428 | ribonuclease inhibitor activity(GO:0008428) |
| 0.0 | 0.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 4.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 2.7 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.1 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004608 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.5 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 3.4 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 5.3 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.7 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.7 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.5 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.0 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.9 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 2.0 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.8 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.4 | 1.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.4 | 4.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.7 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.2 | 1.6 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 1.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 2.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.2 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.1 | 3.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.3 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.3 | GO:1990130 | EGO complex(GO:0034448) Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.5 | GO:0034464 | BBSome(GO:0034464) |
| 0.1 | 1.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 0.2 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.9 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.9 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.7 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 1.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.4 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.5 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |