Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
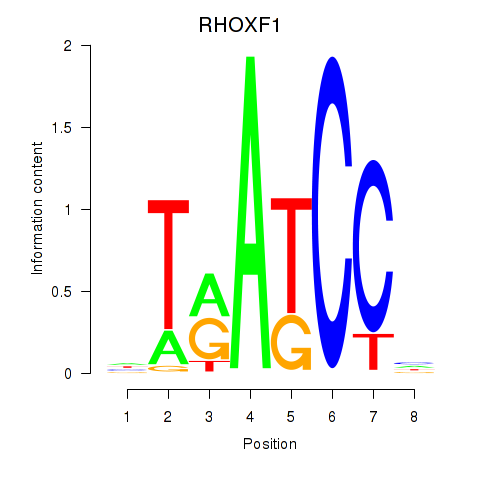
Results for RHOXF1
Z-value: 1.04
Transcription factors associated with RHOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RHOXF1
|
ENSG00000101883.4 | RHOXF1 |
Activity profile of RHOXF1 motif
Sorted Z-values of RHOXF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RHOXF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_54764346 | 3.37 |
ENST00000297313.3 ENST00000344277.6 |
RGS20 |
regulator of G-protein signaling 20 |
| chr1_-_153538011 | 2.71 |
ENST00000368707.4 |
S100A2 |
S100 calcium binding protein A2 |
| chr1_-_151965048 | 2.45 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr2_-_56150910 | 2.23 |
ENST00000424836.2 ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chr4_+_169418195 | 1.98 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr11_+_35211511 | 1.93 |
ENST00000524922.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr1_+_153003671 | 1.89 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr7_+_134464414 | 1.87 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr7_+_55177416 | 1.83 |
ENST00000450046.1 ENST00000454757.2 |
EGFR |
epidermal growth factor receptor |
| chr2_-_56150184 | 1.81 |
ENST00000394554.1 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
| chrX_+_135251783 | 1.81 |
ENST00000394153.2 |
FHL1 |
four and a half LIM domains 1 |
| chr9_+_75766652 | 1.76 |
ENST00000257497.6 |
ANXA1 |
annexin A1 |
| chr18_+_21452804 | 1.76 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr1_-_153363452 | 1.75 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr18_+_61143994 | 1.75 |
ENST00000382771.4 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr7_-_107642348 | 1.72 |
ENST00000393561.1 |
LAMB1 |
laminin, beta 1 |
| chrX_-_10851762 | 1.71 |
ENST00000380785.1 ENST00000380787.1 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
| chr5_+_135385202 | 1.67 |
ENST00000514554.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr9_+_470288 | 1.65 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr11_-_119993979 | 1.59 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr16_+_56598961 | 1.58 |
ENST00000219162.3 |
MT4 |
metallothionein 4 |
| chr4_+_169418255 | 1.52 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr16_+_56691838 | 1.52 |
ENST00000394501.2 |
MT1F |
metallothionein 1F |
| chr6_+_151646800 | 1.50 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr7_+_134464376 | 1.49 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr3_+_122044084 | 1.47 |
ENST00000264474.3 ENST00000479204.1 |
CSTA |
cystatin A (stefin A) |
| chr18_+_21452964 | 1.46 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr6_+_121756809 | 1.44 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr4_+_169842707 | 1.43 |
ENST00000503290.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr3_-_158390282 | 1.43 |
ENST00000264265.3 |
LXN |
latexin |
| chr2_+_192141611 | 1.43 |
ENST00000392316.1 |
MYO1B |
myosin IB |
| chr1_+_81771806 | 1.40 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr16_+_56691911 | 1.40 |
ENST00000568475.1 |
MT1F |
metallothionein 1F |
| chr3_-_189840223 | 1.37 |
ENST00000427335.2 |
LEPREL1 |
leprecan-like 1 |
| chr11_+_35201826 | 1.37 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr7_+_134551583 | 1.36 |
ENST00000435928.1 |
CALD1 |
caldesmon 1 |
| chr16_+_56691606 | 1.34 |
ENST00000334350.6 |
MT1F |
metallothionein 1F |
| chr10_-_126716459 | 1.34 |
ENST00000309035.6 |
CTBP2 |
C-terminal binding protein 2 |
| chr21_-_28217721 | 1.32 |
ENST00000284984.3 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr5_+_135394840 | 1.27 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr2_-_235405168 | 1.27 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr12_-_52887034 | 1.25 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr10_-_105845674 | 1.23 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr11_+_35211429 | 1.15 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr16_-_65155979 | 1.14 |
ENST00000562325.1 ENST00000268603.4 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr6_+_151561506 | 1.10 |
ENST00000253332.1 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr6_+_151561085 | 1.10 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr5_+_66124590 | 1.09 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr9_+_112542572 | 1.05 |
ENST00000374530.3 |
PALM2-AKAP2 |
PALM2-AKAP2 readthrough |
| chr5_+_76114758 | 1.05 |
ENST00000514165.1 ENST00000296677.4 |
F2RL1 |
coagulation factor II (thrombin) receptor-like 1 |
| chr11_+_35198243 | 1.04 |
ENST00000528455.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr3_+_187930719 | 1.04 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_190044480 | 1.04 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr4_-_99578789 | 1.04 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr7_+_134528635 | 1.03 |
ENST00000445569.2 |
CALD1 |
caldesmon 1 |
| chr7_-_94285472 | 1.03 |
ENST00000437425.2 ENST00000447873.1 ENST00000415788.2 |
SGCE |
sarcoglycan, epsilon |
| chr9_+_116298778 | 1.02 |
ENST00000462143.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr5_-_16936340 | 1.02 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr11_-_119993734 | 1.02 |
ENST00000533302.1 |
TRIM29 |
tripartite motif containing 29 |
| chr22_+_31488433 | 1.00 |
ENST00000455608.1 |
SMTN |
smoothelin |
| chr7_-_94285511 | 1.00 |
ENST00000265735.7 |
SGCE |
sarcoglycan, epsilon |
| chr7_-_94285402 | 0.99 |
ENST00000428696.2 ENST00000445866.2 |
SGCE |
sarcoglycan, epsilon |
| chr11_+_35198118 | 0.95 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr9_-_77703115 | 0.90 |
ENST00000361092.4 ENST00000376808.4 |
NMRK1 |
nicotinamide riboside kinase 1 |
| chr7_+_134576317 | 0.89 |
ENST00000424922.1 ENST00000495522.1 |
CALD1 |
caldesmon 1 |
| chr1_+_27189631 | 0.88 |
ENST00000339276.4 |
SFN |
stratifin |
| chr4_-_99578776 | 0.87 |
ENST00000515287.1 |
TSPAN5 |
tetraspanin 5 |
| chr1_+_169079823 | 0.87 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chrX_+_152082969 | 0.86 |
ENST00000535861.1 ENST00000539731.1 ENST00000449285.2 ENST00000318504.7 ENST00000324823.6 ENST00000370268.4 ENST00000370270.2 |
ZNF185 |
zinc finger protein 185 (LIM domain) |
| chr13_+_43597269 | 0.85 |
ENST00000379221.2 |
DNAJC15 |
DnaJ (Hsp40) homolog, subfamily C, member 15 |
| chr12_-_91546926 | 0.85 |
ENST00000550758.1 |
DCN |
decorin |
| chr3_+_158787041 | 0.84 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr3_-_149095652 | 0.84 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr19_+_48216600 | 0.83 |
ENST00000263277.3 ENST00000538399.1 |
EHD2 |
EH-domain containing 2 |
| chr11_-_62314268 | 0.83 |
ENST00000257247.7 ENST00000531324.1 ENST00000378024.4 |
AHNAK |
AHNAK nucleoprotein |
| chr16_+_31483374 | 0.82 |
ENST00000394863.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chrX_+_135251835 | 0.81 |
ENST00000456445.1 |
FHL1 |
four and a half LIM domains 1 |
| chr5_+_140734570 | 0.81 |
ENST00000571252.1 |
PCDHGA4 |
protocadherin gamma subfamily A, 4 |
| chr13_+_73632897 | 0.80 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr1_-_207206092 | 0.80 |
ENST00000359470.5 ENST00000461135.2 |
C1orf116 |
chromosome 1 open reading frame 116 |
| chr1_-_150780757 | 0.79 |
ENST00000271651.3 |
CTSK |
cathepsin K |
| chr3_+_38017264 | 0.77 |
ENST00000436654.1 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr18_-_21852143 | 0.77 |
ENST00000399443.3 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr19_-_39264072 | 0.76 |
ENST00000599035.1 ENST00000378626.4 |
LGALS7 |
lectin, galactoside-binding, soluble, 7 |
| chrX_+_135279179 | 0.76 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr8_+_22436248 | 0.75 |
ENST00000308354.7 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr22_-_36220420 | 0.75 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr9_+_112542591 | 0.73 |
ENST00000483909.1 ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2 PALM2-AKAP2 AKAP2 |
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr3_-_16524357 | 0.73 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr7_+_76026832 | 0.72 |
ENST00000336517.4 |
ZP3 |
zona pellucida glycoprotein 3 (sperm receptor) |
| chrX_+_135252050 | 0.72 |
ENST00000449474.1 ENST00000345434.3 |
FHL1 |
four and a half LIM domains 1 |
| chr21_-_27542972 | 0.72 |
ENST00000346798.3 ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP |
amyloid beta (A4) precursor protein |
| chr11_+_5710919 | 0.71 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr12_+_53693812 | 0.71 |
ENST00000549488.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr22_-_45608324 | 0.70 |
ENST00000496226.1 ENST00000251993.7 |
KIAA0930 |
KIAA0930 |
| chrX_+_51928002 | 0.70 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr16_-_65155833 | 0.69 |
ENST00000566827.1 ENST00000394156.3 ENST00000562998.1 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
| chr8_+_22436635 | 0.68 |
ENST00000452226.1 ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr14_-_61747949 | 0.66 |
ENST00000355702.2 |
TMEM30B |
transmembrane protein 30B |
| chr9_+_17134980 | 0.66 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr9_+_116207007 | 0.66 |
ENST00000374140.2 |
RGS3 |
regulator of G-protein signaling 3 |
| chr16_-_10674528 | 0.65 |
ENST00000359543.3 |
EMP2 |
epithelial membrane protein 2 |
| chr1_+_156030937 | 0.65 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chrX_-_48931648 | 0.64 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr10_-_33623826 | 0.64 |
ENST00000374867.2 |
NRP1 |
neuropilin 1 |
| chr15_+_25200074 | 0.63 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr3_-_133969673 | 0.63 |
ENST00000427044.2 |
RYK |
receptor-like tyrosine kinase |
| chr5_+_140753444 | 0.63 |
ENST00000517434.1 |
PCDHGA6 |
protocadherin gamma subfamily A, 6 |
| chr20_-_17641097 | 0.61 |
ENST00000246043.4 |
RRBP1 |
ribosome binding protein 1 |
| chr5_-_54830871 | 0.59 |
ENST00000307259.8 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
| chr3_+_36421826 | 0.59 |
ENST00000273183.3 |
STAC |
SH3 and cysteine rich domain |
| chr18_+_29027696 | 0.57 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr2_+_33172012 | 0.57 |
ENST00000404816.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr11_-_119991589 | 0.57 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr7_+_90338712 | 0.56 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr15_+_25200108 | 0.56 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr11_+_66276550 | 0.56 |
ENST00000419755.3 |
CTD-3074O7.11 |
Bardet-Biedl syndrome 1 protein |
| chr8_-_17533838 | 0.55 |
ENST00000400046.1 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr8_-_27462822 | 0.55 |
ENST00000522098.1 |
CLU |
clusterin |
| chr1_+_163038565 | 0.55 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr19_-_36643329 | 0.54 |
ENST00000589154.1 |
COX7A1 |
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr14_-_70883708 | 0.54 |
ENST00000256366.4 |
SYNJ2BP |
synaptojanin 2 binding protein |
| chr14_-_24768913 | 0.54 |
ENST00000288111.7 |
DHRS1 |
dehydrogenase/reductase (SDR family) member 1 |
| chr1_-_201368653 | 0.54 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr1_+_169077172 | 0.53 |
ENST00000499679.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr14_-_67826486 | 0.52 |
ENST00000555431.1 ENST00000554236.1 ENST00000555474.1 |
ATP6V1D |
ATPase, H+ transporting, lysosomal 34kDa, V1 subunit D |
| chr5_-_111312622 | 0.52 |
ENST00000395634.3 |
NREP |
neuronal regeneration related protein |
| chr1_-_153599426 | 0.52 |
ENST00000392622.1 |
S100A13 |
S100 calcium binding protein A13 |
| chr11_-_102651343 | 0.52 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr18_-_47813940 | 0.52 |
ENST00000586837.1 ENST00000412036.2 ENST00000589940.1 |
CXXC1 |
CXXC finger protein 1 |
| chr5_+_35856951 | 0.51 |
ENST00000303115.3 ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R |
interleukin 7 receptor |
| chrX_+_107069063 | 0.51 |
ENST00000262843.6 |
MID2 |
midline 2 |
| chr12_-_9268707 | 0.51 |
ENST00000318602.7 |
A2M |
alpha-2-macroglobulin |
| chr8_-_18666360 | 0.50 |
ENST00000286485.8 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr15_-_52483566 | 0.50 |
ENST00000261837.7 |
GNB5 |
guanine nucleotide binding protein (G protein), beta 5 |
| chr11_+_12766583 | 0.50 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr11_+_32112431 | 0.49 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr17_-_18161870 | 0.49 |
ENST00000579294.1 ENST00000545457.2 ENST00000379450.4 ENST00000578558.1 |
FLII |
flightless I homolog (Drosophila) |
| chr12_-_104443890 | 0.49 |
ENST00000547583.1 ENST00000360814.4 ENST00000546851.1 |
GLT8D2 |
glycosyltransferase 8 domain containing 2 |
| chr12_-_91539918 | 0.49 |
ENST00000548218.1 |
DCN |
decorin |
| chr9_-_77703056 | 0.48 |
ENST00000376811.1 |
NMRK1 |
nicotinamide riboside kinase 1 |
| chr1_-_146082633 | 0.48 |
ENST00000605317.1 ENST00000604938.1 ENST00000339388.5 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chrX_-_151999269 | 0.48 |
ENST00000370277.3 |
CETN2 |
centrin, EF-hand protein, 2 |
| chr1_+_40506392 | 0.47 |
ENST00000414893.1 ENST00000414281.1 ENST00000420216.1 ENST00000372792.2 ENST00000372798.1 ENST00000340450.3 ENST00000372805.3 ENST00000435719.1 ENST00000427843.1 ENST00000417287.1 ENST00000424977.1 ENST00000446031.1 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr1_-_201368707 | 0.47 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr9_+_116263639 | 0.47 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr3_-_122134882 | 0.46 |
ENST00000330689.4 |
WDR5B |
WD repeat domain 5B |
| chr3_-_121468513 | 0.46 |
ENST00000494517.1 ENST00000393667.3 |
GOLGB1 |
golgin B1 |
| chr9_-_13279563 | 0.46 |
ENST00000541718.1 |
MPDZ |
multiple PDZ domain protein |
| chr2_+_102721023 | 0.45 |
ENST00000409589.1 ENST00000409329.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr19_+_35168633 | 0.45 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chr9_+_116263778 | 0.45 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr9_+_17135016 | 0.45 |
ENST00000425824.1 ENST00000262360.5 ENST00000380641.4 |
CNTLN |
centlein, centrosomal protein |
| chr9_-_95298314 | 0.44 |
ENST00000344604.5 ENST00000375540.1 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
| chr5_+_140797296 | 0.44 |
ENST00000398594.2 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
| chr11_-_76381029 | 0.43 |
ENST00000407242.2 ENST00000421973.1 |
LRRC32 |
leucine rich repeat containing 32 |
| chr5_+_95997769 | 0.43 |
ENST00000338252.3 ENST00000508830.1 |
CAST |
calpastatin |
| chr3_-_133969437 | 0.42 |
ENST00000460933.1 ENST00000296084.4 |
RYK |
receptor-like tyrosine kinase |
| chrX_-_114253536 | 0.42 |
ENST00000371936.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chr6_-_30181156 | 0.42 |
ENST00000418026.1 ENST00000416596.1 ENST00000453195.1 |
TRIM26 |
tripartite motif containing 26 |
| chr3_-_121468602 | 0.42 |
ENST00000340645.5 |
GOLGB1 |
golgin B1 |
| chr16_+_8806800 | 0.41 |
ENST00000561870.1 ENST00000396600.2 |
ABAT |
4-aminobutyrate aminotransferase |
| chr8_+_9911778 | 0.40 |
ENST00000317173.4 ENST00000441698.2 |
MSRA |
methionine sulfoxide reductase A |
| chr1_-_45140227 | 0.40 |
ENST00000372237.3 |
TMEM53 |
transmembrane protein 53 |
| chr17_+_65374075 | 0.40 |
ENST00000581322.1 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr11_+_102980126 | 0.39 |
ENST00000375735.2 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr6_-_32820529 | 0.39 |
ENST00000425148.2 |
TAP1 |
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr17_-_1395954 | 0.39 |
ENST00000359786.5 |
MYO1C |
myosin IC |
| chr1_+_15479054 | 0.39 |
ENST00000376014.3 ENST00000451326.2 |
TMEM51 |
transmembrane protein 51 |
| chr18_-_28682374 | 0.39 |
ENST00000280904.6 |
DSC2 |
desmocollin 2 |
| chr7_-_76256557 | 0.39 |
ENST00000275569.4 ENST00000310842.4 |
POMZP3 |
POM121 and ZP3 fusion |
| chr2_-_175711133 | 0.39 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr7_+_23286182 | 0.38 |
ENST00000258733.4 ENST00000381990.2 ENST00000409458.3 ENST00000539136.1 ENST00000453162.2 |
GPNMB |
glycoprotein (transmembrane) nmb |
| chr16_-_3073933 | 0.38 |
ENST00000574151.1 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
| chr7_+_128379346 | 0.38 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr12_-_110888103 | 0.37 |
ENST00000426440.1 ENST00000228825.7 |
ARPC3 |
actin related protein 2/3 complex, subunit 3, 21kDa |
| chr11_+_121461097 | 0.37 |
ENST00000527934.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr9_-_112083229 | 0.37 |
ENST00000374566.3 ENST00000374557.4 |
EPB41L4B |
erythrocyte membrane protein band 4.1 like 4B |
| chr12_+_53693466 | 0.37 |
ENST00000267103.5 ENST00000548632.1 |
C12orf10 |
chromosome 12 open reading frame 10 |
| chr20_+_6748311 | 0.36 |
ENST00000378827.4 |
BMP2 |
bone morphogenetic protein 2 |
| chr11_-_67120974 | 0.36 |
ENST00000539074.1 ENST00000312419.3 |
POLD4 |
polymerase (DNA-directed), delta 4, accessory subunit |
| chr20_+_33759854 | 0.36 |
ENST00000216968.4 |
PROCR |
protein C receptor, endothelial |
| chr12_-_56121612 | 0.36 |
ENST00000546939.1 |
CD63 |
CD63 molecule |
| chr6_+_35310312 | 0.35 |
ENST00000448077.2 ENST00000360694.3 ENST00000418635.2 ENST00000444397.1 |
PPARD |
peroxisome proliferator-activated receptor delta |
| chr8_+_22428457 | 0.35 |
ENST00000517962.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr8_-_27468842 | 0.35 |
ENST00000523500.1 |
CLU |
clusterin |
| chr6_+_110012462 | 0.35 |
ENST00000441478.2 ENST00000230124.3 |
FIG4 |
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
| chr6_+_90272027 | 0.35 |
ENST00000522441.1 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr9_-_13279589 | 0.34 |
ENST00000319217.7 |
MPDZ |
multiple PDZ domain protein |
| chr8_-_27468945 | 0.34 |
ENST00000405140.3 |
CLU |
clusterin |
| chrX_+_99899180 | 0.34 |
ENST00000373004.3 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
| chr5_+_140593509 | 0.34 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chrX_-_8700171 | 0.33 |
ENST00000262648.3 |
KAL1 |
Kallmann syndrome 1 sequence |
| chr18_+_11752040 | 0.33 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr3_+_111718036 | 0.33 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr17_+_4843679 | 0.33 |
ENST00000576229.1 |
RNF167 |
ring finger protein 167 |
| chr16_-_18430593 | 0.33 |
ENST00000525596.1 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
| chr17_+_74381343 | 0.33 |
ENST00000392496.3 |
SPHK1 |
sphingosine kinase 1 |
| chr1_-_45140074 | 0.33 |
ENST00000420706.1 ENST00000372235.3 ENST00000372242.3 ENST00000372243.3 ENST00000372244.3 |
TMEM53 |
transmembrane protein 53 |
| chr4_+_88754113 | 0.33 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.6 | 1.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 1.4 | GO:0061769 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.3 | 1.6 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.3 | 0.9 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.3 | 1.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 6.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 3.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 6.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.2 | 4.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.2 | 1.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.2 | 1.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.2 | 0.5 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.2 | 1.0 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 3.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 3.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.1 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.1 | 1.0 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.5 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 1.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.8 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.1 | 0.3 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.1 | 1.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.5 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.4 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.1 | 0.4 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.7 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.2 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 0.8 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 0.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.1 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 0.3 | GO:1901474 | azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 1.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 4.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.7 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 0.3 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.6 | GO:0031432 | titin binding(GO:0031432) |
| 0.1 | 0.5 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.3 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.1 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.2 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.2 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.4 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0098625 | methylselenol reductase activity(GO:0098625) methylseleninic acid reductase activity(GO:0098626) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.1 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0030883 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.8 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 6.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.0 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.4 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0070679 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 1.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.0 | 2.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.0 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.6 | 2.5 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.6 | 1.8 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.6 | 1.8 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.5 | 1.6 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.4 | 4.0 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.4 | 1.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 1.3 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 1.4 | GO:1903288 | protein transport into plasma membrane raft(GO:0044861) positive regulation of potassium ion import(GO:1903288) |
| 0.3 | 0.8 | GO:2000196 | positive regulation of female gonad development(GO:2000196) |
| 0.3 | 1.1 | GO:0061643 | chemorepulsion of dopaminergic neuron axon(GO:0036518) chemorepulsion of axon(GO:0061643) |
| 0.3 | 1.6 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.3 | 1.0 | GO:1903224 | regulation of endodermal cell differentiation(GO:1903224) |
| 0.3 | 3.9 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.3 | 1.8 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.2 | 5.7 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.7 | GO:0032594 | protein transport within lipid bilayer(GO:0032594) protein transport into membrane raft(GO:0032596) |
| 0.2 | 1.0 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) endothelial tip cell fate specification(GO:0097102) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.2 | 1.8 | GO:0021957 | corticospinal tract morphogenesis(GO:0021957) |
| 0.2 | 0.4 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.2 | 3.1 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 1.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.2 | 2.2 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.2 | 0.8 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 4.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 0.3 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.2 | 0.5 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 1.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 4.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.4 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.6 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.1 | 0.9 | GO:1902957 | negative regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902957) |
| 0.1 | 0.7 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.5 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.2 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.1 | 0.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.1 | GO:1900104 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.3 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.9 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.1 | 0.6 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 0.4 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.3 | GO:0070428 | negative regulation of interleukin-23 production(GO:0032707) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.1 | 0.4 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.1 | 1.3 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.3 | GO:0086100 | endothelin receptor signaling pathway(GO:0086100) |
| 0.1 | 1.2 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.1 | 0.3 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.3 | GO:0018874 | benzoate metabolic process(GO:0018874) butyrate metabolic process(GO:0019605) |
| 0.1 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.1 | 0.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.7 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 3.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.4 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.1 | 0.1 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 2.7 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 3.8 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.1 | 0.3 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.2 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.2 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.1 | 0.2 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.7 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.1 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.1 | GO:0060796 | regulation of transcription involved in primary germ layer cell fate commitment(GO:0060796) |
| 0.1 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.3 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.5 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.5 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.1 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.1 | 0.5 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 0.4 | GO:0043416 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.1 | 0.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.1 | 0.1 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 0.3 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.1 | 0.2 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.2 | GO:0044501 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 1.3 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 | 0.3 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.4 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.1 | 0.3 | GO:0033384 | geranyl diphosphate metabolic process(GO:0033383) geranyl diphosphate biosynthetic process(GO:0033384) farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.2 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.1 | 0.5 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.3 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.1 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.2 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.2 | GO:0002331 | pre-B cell differentiation(GO:0002329) pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.7 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.2 | GO:1901098 | regulation of autophagosome maturation(GO:1901096) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.2 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.4 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.5 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.5 | GO:0033089 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 1.4 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.1 | GO:2000547 | dendritic cell dendrite assembly(GO:0097026) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.4 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.0 | 1.7 | GO:0060512 | prostate gland morphogenesis(GO:0060512) |
| 0.0 | 0.3 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.0 | 0.5 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 1.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:1902462 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.2 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) positive regulation of transforming growth factor beta3 production(GO:0032916) negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.2 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.5 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0048200 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0015803 | branched-chain amino acid transport(GO:0015803) |
| 0.0 | 0.2 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 3.3 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.4 | GO:0001649 | osteoblast differentiation(GO:0001649) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 1.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.1 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.1 | GO:0086053 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.2 | GO:0016045 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.1 | GO:0022029 | cerebral cortex cell migration(GO:0021795) forebrain cell migration(GO:0021885) telencephalon cell migration(GO:0022029) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.1 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0061535 | glutamate secretion, neurotransmission(GO:0061535) |
| 0.0 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.3 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.2 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.4 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.7 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:1901079 | dense core granule biogenesis(GO:0061110) positive regulation of relaxation of muscle(GO:1901079) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.0 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 3.1 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.0 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.3 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.0 | 0.1 | GO:1900369 | regulation of RNA interference(GO:1900368) negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.6 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:2001191 | gamma-delta T cell activation involved in immune response(GO:0002290) negative regulation of interferon-beta secretion(GO:0035548) regulation of gamma-delta T cell activation involved in immune response(GO:2001191) positive regulation of gamma-delta T cell activation involved in immune response(GO:2001193) regulation of transforming growth factor-beta secretion(GO:2001201) negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.2 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.5 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.9 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.2 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0014040 | positive regulation of Schwann cell differentiation(GO:0014040) |
| 0.0 | 0.1 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0014902 | myotube differentiation(GO:0014902) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.1 | GO:0010286 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.3 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 2.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.0 | 0.2 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.3 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 0.1 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0022604 | regulation of cell morphogenesis(GO:0022604) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.2 | GO:0045986 | negative regulation of smooth muscle contraction(GO:0045986) |
| 0.0 | 0.1 | GO:2000674 | regulation of type B pancreatic cell apoptotic process(GO:2000674) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.0 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.4 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.0 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.4 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.3 | GO:0050704 | regulation of interleukin-1 secretion(GO:0050704) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.0 | GO:0000965 | mitochondrial RNA 3'-end processing(GO:0000965) |
| 0.0 | 0.1 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.2 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.0 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.5 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 1.8 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0051902 | negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.0 | 0.4 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.1 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 2.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.1 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.4 | GO:0006266 | DNA ligation(GO:0006266) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:1990090 | cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.0 | GO:0033087 | regulation of immature T cell proliferation(GO:0033083) negative regulation of immature T cell proliferation(GO:0033087) |
| 0.0 | 0.2 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 1.8 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.3 | GO:0015800 | acidic amino acid transport(GO:0015800) L-glutamate transport(GO:0015813) |
| 0.0 | 0.8 | GO:0006939 | smooth muscle contraction(GO:0006939) |
| 0.0 | 0.1 | GO:0071600 | otic vesicle formation(GO:0030916) otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.3 | GO:0050912 | detection of chemical stimulus involved in sensory perception of taste(GO:0050912) |
| 0.0 | 0.0 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.2 | GO:0051194 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.2 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.4 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.6 | 1.8 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.6 | 1.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 6.6 | GO:0030478 | actin cap(GO:0030478) |
| 0.4 | 3.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 3.0 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.3 | 1.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.3 | 1.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.7 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.2 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 1.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 5.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.3 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.4 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.1 | 0.5 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 1.6 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 1.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.4 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 1.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) sorting endosome(GO:0097443) |
| 0.1 | 0.7 | GO:1990812 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.1 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 1.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 1.1 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 1.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.2 | GO:0071664 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.3 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 4.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.8 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.3 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 1.0 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.9 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.7 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.5 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 3.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 1.6 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0036021 | endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 11.4 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.6 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 2.1 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.2 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 6.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 4.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 2.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 3.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 3.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 8.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.4 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 3.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.0 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 7.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.3 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 4.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 1.8 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 1.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.3 | REACTOME SIGNALING BY WNT | Genes involved in Signaling by Wnt |
| 0.1 | 1.0 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 1.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 3.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 3.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 1.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 1.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 3.2 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 2.6 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |