Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
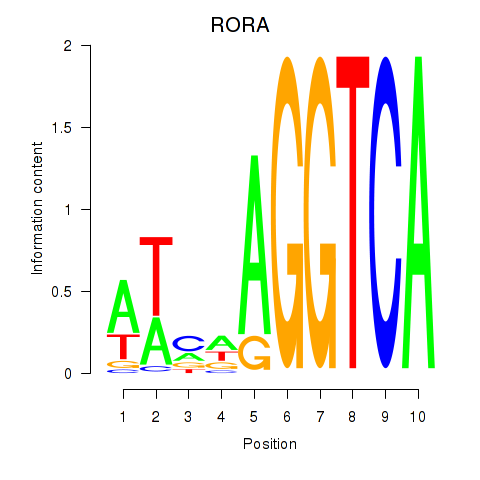
Results for RORA
Z-value: 0.90
Transcription factors associated with RORA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORA
|
ENSG00000069667.11 | RORA |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RORA | hg19_v2_chr15_-_60884706_60884743, hg19_v2_chr15_-_61521495_61521518 | -0.08 | 7.6e-01 | Click! |
Activity profile of RORA motif
Sorted Z-values of RORA motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RORA
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_93964158 | 3.33 |
ENST00000549206.1 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr12_+_93963590 | 2.95 |
ENST00000340600.2 |
SOCS2 |
suppressor of cytokine signaling 2 |
| chr16_-_88717423 | 2.00 |
ENST00000568278.1 ENST00000569359.1 ENST00000567174.1 |
CYBA |
cytochrome b-245, alpha polypeptide |
| chr7_+_112063192 | 1.36 |
ENST00000005558.4 |
IFRD1 |
interferon-related developmental regulator 1 |
| chr1_-_85156216 | 1.34 |
ENST00000342203.3 ENST00000370612.4 |
SSX2IP |
synovial sarcoma, X breakpoint 2 interacting protein |
| chr3_-_141747950 | 1.22 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr6_-_41039567 | 1.19 |
ENST00000468811.1 |
OARD1 |
O-acyl-ADP-ribose deacylase 1 |
| chr9_-_130966497 | 1.17 |
ENST00000393608.1 ENST00000372948.3 |
CIZ1 |
CDKN1A interacting zinc finger protein 1 |
| chrX_-_71526999 | 1.10 |
ENST00000453707.2 ENST00000373619.3 |
CITED1 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr7_-_45151272 | 1.07 |
ENST00000461363.1 ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4 |
transforming growth factor beta regulator 4 |
| chr7_-_123197733 | 1.00 |
ENST00000470123.1 ENST00000471770.1 |
NDUFA5 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr10_+_81107271 | 0.98 |
ENST00000448165.1 |
PPIF |
peptidylprolyl isomerase F |
| chr9_+_134103496 | 0.87 |
ENST00000498010.1 ENST00000476004.1 ENST00000528406.1 |
NUP214 |
nucleoporin 214kDa |
| chr1_-_150207017 | 0.81 |
ENST00000369119.3 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_-_48785216 | 0.76 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr11_-_64511575 | 0.72 |
ENST00000431822.1 ENST00000377486.3 ENST00000394432.3 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr19_-_55669093 | 0.71 |
ENST00000344887.5 |
TNNI3 |
troponin I type 3 (cardiac) |
| chr1_+_206858328 | 0.68 |
ENST00000367103.3 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
| chr1_+_229440129 | 0.65 |
ENST00000366688.3 |
SPHAR |
S-phase response (cyclin related) |
| chr21_-_43816152 | 0.58 |
ENST00000433957.2 ENST00000398397.3 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chr1_-_32403903 | 0.57 |
ENST00000344035.6 ENST00000356536.3 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
| chrX_-_43741594 | 0.56 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr1_+_212738676 | 0.55 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr21_-_43816052 | 0.55 |
ENST00000398405.1 |
TMPRSS3 |
transmembrane protease, serine 3 |
| chrX_-_129299638 | 0.55 |
ENST00000535724.1 ENST00000346424.2 |
AIFM1 |
apoptosis-inducing factor, mitochondrion-associated, 1 |
| chr18_-_19284724 | 0.50 |
ENST00000580981.1 ENST00000289119.2 |
ABHD3 |
abhydrolase domain containing 3 |
| chr1_-_33502441 | 0.50 |
ENST00000548033.1 ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2 |
adenylate kinase 2 |
| chr11_-_64512273 | 0.48 |
ENST00000377497.3 ENST00000377487.1 ENST00000430645.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr17_-_41132010 | 0.48 |
ENST00000409103.1 ENST00000360221.4 |
PTGES3L-AARSD1 |
PTGES3L-AARSD1 readthrough |
| chr19_-_39826639 | 0.48 |
ENST00000602185.1 ENST00000598034.1 ENST00000601387.1 ENST00000595636.1 ENST00000253054.8 ENST00000594700.1 ENST00000597595.1 |
GMFG |
glia maturation factor, gamma |
| chr19_-_50311896 | 0.44 |
ENST00000529634.2 |
FUZ |
fuzzy planar cell polarity protein |
| chr1_+_206858232 | 0.44 |
ENST00000294981.4 |
MAPKAPK2 |
mitogen-activated protein kinase-activated protein kinase 2 |
| chr11_-_104480019 | 0.43 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chr6_-_41703296 | 0.43 |
ENST00000373033.1 |
TFEB |
transcription factor EB |
| chr11_+_64879317 | 0.42 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr1_-_33502528 | 0.42 |
ENST00000354858.6 |
AK2 |
adenylate kinase 2 |
| chr18_-_12377283 | 0.42 |
ENST00000269143.3 |
AFG3L2 |
AFG3-like AAA ATPase 2 |
| chr11_+_111957497 | 0.42 |
ENST00000375549.3 ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD |
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr16_-_30457048 | 0.42 |
ENST00000500504.2 ENST00000542752.1 |
SEPHS2 |
selenophosphate synthetase 2 |
| chr1_-_241799232 | 0.42 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr14_-_102552659 | 0.41 |
ENST00000441629.2 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr7_-_140179276 | 0.40 |
ENST00000443720.2 ENST00000255977.2 |
MKRN1 |
makorin ring finger protein 1 |
| chr3_-_24536253 | 0.40 |
ENST00000428492.1 ENST00000396671.2 ENST00000431815.1 ENST00000418247.1 ENST00000416420.1 ENST00000356447.4 |
THRB |
thyroid hormone receptor, beta |
| chr19_+_46850251 | 0.40 |
ENST00000012443.4 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr12_+_120875910 | 0.39 |
ENST00000551806.1 |
AL021546.6 |
Glutamyl-tRNA(Gln) amidotransferase subunit C, mitochondrial |
| chr12_-_53320245 | 0.39 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chr16_-_30538079 | 0.38 |
ENST00000562803.1 |
ZNF768 |
zinc finger protein 768 |
| chr10_-_52645379 | 0.37 |
ENST00000395489.2 |
A1CF |
APOBEC1 complementation factor |
| chr19_+_46850320 | 0.36 |
ENST00000391919.1 |
PPP5C |
protein phosphatase 5, catalytic subunit |
| chr17_+_49230897 | 0.36 |
ENST00000393196.3 ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1 NME1-NME2 NME2 |
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chr11_+_66624527 | 0.35 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr16_-_30537839 | 0.35 |
ENST00000380412.5 |
ZNF768 |
zinc finger protein 768 |
| chr7_-_127983877 | 0.35 |
ENST00000415472.2 ENST00000478061.1 ENST00000223073.2 ENST00000459726.1 |
RBM28 |
RNA binding motif protein 28 |
| chr10_-_52645416 | 0.34 |
ENST00000374001.2 ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF |
APOBEC1 complementation factor |
| chr6_-_89673280 | 0.34 |
ENST00000369475.3 ENST00000369485.4 ENST00000538899.1 ENST00000265607.6 |
RNGTT |
RNA guanylyltransferase and 5'-phosphatase |
| chr7_-_25164969 | 0.34 |
ENST00000305786.2 |
CYCS |
cytochrome c, somatic |
| chr1_+_32666188 | 0.33 |
ENST00000421922.2 |
CCDC28B |
coiled-coil domain containing 28B |
| chr3_+_113465866 | 0.32 |
ENST00000273398.3 ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A |
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr6_-_97345689 | 0.31 |
ENST00000316149.7 |
NDUFAF4 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr10_-_104178857 | 0.30 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr6_+_107077435 | 0.30 |
ENST00000369046.4 |
QRSL1 |
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr19_+_16059818 | 0.30 |
ENST00000322107.1 |
OR10H4 |
olfactory receptor, family 10, subfamily H, member 4 |
| chr17_-_42144949 | 0.30 |
ENST00000591247.1 |
LSM12 |
LSM12 homolog (S. cerevisiae) |
| chr16_+_21964662 | 0.29 |
ENST00000561553.1 ENST00000565331.1 |
UQCRC2 |
ubiquinol-cytochrome c reductase core protein II |
| chr6_+_167525277 | 0.28 |
ENST00000400926.2 |
CCR6 |
chemokine (C-C motif) receptor 6 |
| chr21_-_46330545 | 0.28 |
ENST00000320216.6 ENST00000397852.1 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
| chr9_+_139717847 | 0.27 |
ENST00000436380.1 |
RABL6 |
RAB, member RAS oncogene family-like 6 |
| chr3_-_183735731 | 0.27 |
ENST00000334444.6 |
ABCC5 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr15_+_78441663 | 0.27 |
ENST00000299518.2 ENST00000558554.1 ENST00000557826.1 ENST00000561279.1 ENST00000559186.1 ENST00000560770.1 ENST00000559881.1 ENST00000559205.1 ENST00000441490.2 |
IDH3A |
isocitrate dehydrogenase 3 (NAD+) alpha |
| chr7_+_73868439 | 0.26 |
ENST00000424337.2 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
| chr9_-_35103105 | 0.25 |
ENST00000452248.2 ENST00000356493.5 |
STOML2 |
stomatin (EPB72)-like 2 |
| chr15_+_43809797 | 0.25 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr5_-_140027357 | 0.25 |
ENST00000252102.4 |
NDUFA2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr5_-_140027175 | 0.24 |
ENST00000512088.1 |
NDUFA2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa |
| chr11_-_67271723 | 0.23 |
ENST00000533391.1 ENST00000534749.1 ENST00000532703.1 |
PITPNM1 |
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr16_-_1821496 | 0.22 |
ENST00000564628.1 ENST00000563498.1 |
NME3 |
NME/NM23 nucleoside diphosphate kinase 3 |
| chr5_+_169780485 | 0.22 |
ENST00000377360.4 |
KCNIP1 |
Kv channel interacting protein 1 |
| chr2_+_198365095 | 0.22 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr10_-_94003003 | 0.22 |
ENST00000412050.4 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
| chr19_-_11689752 | 0.21 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr1_-_85040090 | 0.20 |
ENST00000370630.5 |
CTBS |
chitobiase, di-N-acetyl- |
| chr14_-_101295407 | 0.20 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chr3_+_45067659 | 0.20 |
ENST00000296130.4 |
CLEC3B |
C-type lectin domain family 3, member B |
| chr11_+_118272328 | 0.19 |
ENST00000524422.1 |
ATP5L |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit G |
| chr11_-_111741994 | 0.19 |
ENST00000398006.2 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
| chr3_-_113465065 | 0.19 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr17_+_40996590 | 0.18 |
ENST00000253799.3 ENST00000452774.2 |
AOC2 |
amine oxidase, copper containing 2 (retina-specific) |
| chr11_-_132813566 | 0.17 |
ENST00000331898.7 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
| chr18_+_32073253 | 0.16 |
ENST00000283365.9 ENST00000596745.1 ENST00000315456.6 |
DTNA |
dystrobrevin, alpha |
| chr7_-_150777874 | 0.16 |
ENST00000540185.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr19_-_39926268 | 0.15 |
ENST00000599705.1 |
RPS16 |
ribosomal protein S16 |
| chr10_+_104178946 | 0.15 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr16_+_72088376 | 0.14 |
ENST00000570083.1 ENST00000355906.5 ENST00000398131.2 ENST00000569639.1 ENST00000564499.1 ENST00000357763.4 ENST00000562526.1 ENST00000565574.1 ENST00000568417.2 ENST00000356967.5 |
HP HPR |
haptoglobin haptoglobin-related protein |
| chr20_-_3748416 | 0.14 |
ENST00000399672.1 |
C20orf27 |
chromosome 20 open reading frame 27 |
| chr14_-_58893832 | 0.14 |
ENST00000556007.2 |
TIMM9 |
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr19_-_49140609 | 0.13 |
ENST00000601104.1 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
| chr1_+_29213678 | 0.13 |
ENST00000347529.3 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr11_-_790060 | 0.12 |
ENST00000330106.4 |
CEND1 |
cell cycle exit and neuronal differentiation 1 |
| chr16_+_31366536 | 0.12 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr1_-_205719295 | 0.12 |
ENST00000367142.4 |
NUCKS1 |
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr5_-_131347501 | 0.12 |
ENST00000543479.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr11_-_64511789 | 0.11 |
ENST00000419843.1 ENST00000394430.1 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr17_+_48046538 | 0.10 |
ENST00000240306.3 |
DLX4 |
distal-less homeobox 4 |
| chr3_-_49158312 | 0.10 |
ENST00000398892.3 ENST00000453664.1 ENST00000398888.2 |
USP19 |
ubiquitin specific peptidase 19 |
| chr17_-_49198216 | 0.10 |
ENST00000262013.7 ENST00000357122.4 |
SPAG9 |
sperm associated antigen 9 |
| chr20_+_49126881 | 0.10 |
ENST00000371621.3 ENST00000541713.1 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
| chrX_+_18725758 | 0.10 |
ENST00000472826.1 ENST00000544635.1 ENST00000496075.2 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
| chr12_+_56414851 | 0.09 |
ENST00000547167.1 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
| chr5_-_131347583 | 0.07 |
ENST00000379255.1 ENST00000430403.1 ENST00000544770.1 ENST00000379246.1 ENST00000414078.1 ENST00000441995.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chr3_-_42845951 | 0.07 |
ENST00000418900.2 ENST00000430190.1 |
HIGD1A |
HIG1 hypoxia inducible domain family, member 1A |
| chrX_+_153770421 | 0.07 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr10_-_65028938 | 0.07 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chrX_-_108725301 | 0.07 |
ENST00000218006.2 |
GUCY2F |
guanylate cyclase 2F, retinal |
| chr2_+_162016827 | 0.07 |
ENST00000429217.1 ENST00000406287.1 ENST00000402568.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr19_-_42931567 | 0.07 |
ENST00000244289.4 |
LIPE |
lipase, hormone-sensitive |
| chr11_-_119599794 | 0.07 |
ENST00000264025.3 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chrX_+_16737718 | 0.06 |
ENST00000380155.3 |
SYAP1 |
synapse associated protein 1 |
| chr5_+_32788945 | 0.06 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr16_+_82090028 | 0.05 |
ENST00000568090.1 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr2_-_86422095 | 0.05 |
ENST00000254636.5 |
IMMT |
inner membrane protein, mitochondrial |
| chr16_+_2076869 | 0.05 |
ENST00000424542.2 ENST00000432365.2 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr13_+_22245522 | 0.04 |
ENST00000382353.5 |
FGF9 |
fibroblast growth factor 9 |
| chr20_+_37590942 | 0.04 |
ENST00000373325.2 ENST00000252011.3 ENST00000373323.4 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr12_-_16761007 | 0.04 |
ENST00000354662.1 ENST00000441439.2 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr1_-_154164534 | 0.04 |
ENST00000271850.7 ENST00000368530.2 |
TPM3 |
tropomyosin 3 |
| chr1_+_9294822 | 0.03 |
ENST00000377403.2 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr1_-_157108266 | 0.03 |
ENST00000326786.4 |
ETV3 |
ets variant 3 |
| chr6_-_43021437 | 0.03 |
ENST00000265348.3 |
CUL7 |
cullin 7 |
| chr10_-_14050522 | 0.03 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr1_-_157108130 | 0.02 |
ENST00000368192.4 |
ETV3 |
ets variant 3 |
| chr17_-_27503770 | 0.01 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr8_-_100905850 | 0.01 |
ENST00000520271.1 ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C |
cytochrome c oxidase subunit VIc |
| chr22_-_32341336 | 0.01 |
ENST00000248984.3 |
C22orf24 |
chromosome 22 open reading frame 24 |
| chr12_-_57039739 | 0.01 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 6.3 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.6 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.1 | 0.4 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.1 | 0.4 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.1 | 0.4 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.3 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.3 | GO:0004040 | amidase activity(GO:0004040) |
| 0.1 | 0.5 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 2.0 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.7 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 1.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 1.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.0 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 0.4 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.4 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.0 | 0.2 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 1.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) chitin binding(GO:0008061) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 1.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.2 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.2 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.7 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.2 | GO:0044769 | ATPase activity, coupled to transmembrane movement of ions, rotational mechanism(GO:0044769) proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 1.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 2.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.9 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.2 | 0.7 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.2 | 0.6 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.7 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 2.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.8 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 0.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.1 | 0.4 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.0 | 1.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.8 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.8 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 1.2 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 2.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) apicolateral plasma membrane(GO:0016327) glycoprotein complex(GO:0090665) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0007518 | myoblast fate determination(GO:0007518) |
| 0.4 | 2.0 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.3 | 6.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.3 | 1.0 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.3 | 0.9 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.2 | 1.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.2 | 0.6 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.5 | GO:1904044 | response to aldosterone(GO:1904044) |
| 0.1 | 1.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.4 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.4 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.1 | 0.8 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.1 | 0.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.4 | GO:0090301 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.1 | 1.0 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 0.4 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.3 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 1.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.6 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.2 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.1 | 0.5 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.1 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.3 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.6 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.8 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.4 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.2 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 1.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.3 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.0 | 0.2 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 1.3 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.8 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.3 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 1.3 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.5 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 7.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 1.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 2.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.9 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.7 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |