Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
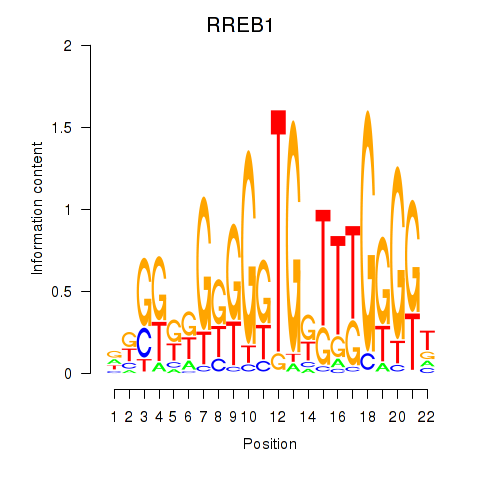
Results for RREB1
Z-value: 1.47
Transcription factors associated with RREB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RREB1
|
ENSG00000124782.15 | RREB1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| RREB1 | hg19_v2_chr6_+_7108210_7108264 | 0.61 | 1.2e-02 | Click! |
Activity profile of RREB1 motif
Sorted Z-values of RREB1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of RREB1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_35609380 | 5.34 |
ENST00000604621.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr1_-_153538292 | 5.33 |
ENST00000497140.1 ENST00000368708.3 |
S100A2 |
S100 calcium binding protein A2 |
| chr11_+_86511569 | 4.06 |
ENST00000441050.1 |
PRSS23 |
protease, serine, 23 |
| chr10_+_17271266 | 3.93 |
ENST00000224237.5 |
VIM |
vimentin |
| chr19_-_51456321 | 3.77 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_-_51456344 | 3.60 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr4_+_74735102 | 3.35 |
ENST00000395761.3 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr17_-_39677971 | 3.11 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr14_-_105635090 | 2.69 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chrX_+_102631844 | 2.64 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr19_-_51456198 | 2.57 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr1_-_153363452 | 2.56 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr8_+_32405785 | 2.36 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr10_-_126849588 | 2.31 |
ENST00000411419.2 |
CTBP2 |
C-terminal binding protein 2 |
| chr2_-_85637459 | 2.21 |
ENST00000409921.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr10_-_126849068 | 2.21 |
ENST00000494626.2 ENST00000337195.5 |
CTBP2 |
C-terminal binding protein 2 |
| chr19_-_19051927 | 1.93 |
ENST00000600077.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr12_+_96588279 | 1.90 |
ENST00000552142.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr9_-_14314066 | 1.85 |
ENST00000397575.3 |
NFIB |
nuclear factor I/B |
| chr17_-_62658186 | 1.78 |
ENST00000262435.9 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
| chr12_+_96588143 | 1.72 |
ENST00000228741.3 ENST00000547249.1 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr9_-_14313893 | 1.72 |
ENST00000380921.3 ENST00000380959.3 |
NFIB |
nuclear factor I/B |
| chr19_-_19051993 | 1.67 |
ENST00000594794.1 ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr4_+_75230853 | 1.67 |
ENST00000244869.2 |
EREG |
epiregulin |
| chr8_+_32406179 | 1.61 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr11_+_69455855 | 1.57 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr7_+_75932863 | 1.56 |
ENST00000429938.1 |
HSPB1 |
heat shock 27kDa protein 1 |
| chrX_+_134166333 | 1.51 |
ENST00000257013.7 |
FAM127A |
family with sequence similarity 127, member A |
| chr12_+_49212514 | 1.40 |
ENST00000301050.2 ENST00000548279.1 ENST00000547230.1 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
| chr5_+_125758865 | 1.27 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr17_+_7788104 | 1.26 |
ENST00000380358.4 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr6_+_7541845 | 1.25 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr5_+_125758813 | 1.23 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr1_-_17307173 | 1.23 |
ENST00000438542.1 ENST00000375535.3 |
MFAP2 |
microfibrillar-associated protein 2 |
| chr19_+_55476620 | 1.21 |
ENST00000543010.1 ENST00000391721.4 ENST00000339757.7 |
NLRP2 |
NLR family, pyrin domain containing 2 |
| chr4_+_56719782 | 1.19 |
ENST00000381295.2 ENST00000346134.7 ENST00000349598.6 |
EXOC1 |
exocyst complex component 1 |
| chr16_+_68771128 | 1.16 |
ENST00000261769.5 ENST00000422392.2 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
| chr19_+_55477711 | 1.16 |
ENST00000448584.2 ENST00000537859.1 ENST00000585500.1 ENST00000427260.2 ENST00000538819.1 ENST00000263437.6 |
NLRP2 |
NLR family, pyrin domain containing 2 |
| chr6_+_7541808 | 1.12 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr9_-_21994597 | 1.10 |
ENST00000579755.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr8_+_94929969 | 1.10 |
ENST00000517764.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr12_-_53074182 | 1.09 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr17_+_35849937 | 1.07 |
ENST00000394389.4 |
DUSP14 |
dual specificity phosphatase 14 |
| chr9_-_21994344 | 1.04 |
ENST00000530628.2 ENST00000361570.3 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr9_+_132934835 | 1.01 |
ENST00000372398.3 |
NCS1 |
neuronal calcium sensor 1 |
| chr9_-_13279563 | 1.01 |
ENST00000541718.1 |
MPDZ |
multiple PDZ domain protein |
| chr22_-_36357671 | 0.99 |
ENST00000408983.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr16_+_27325202 | 0.98 |
ENST00000395762.2 ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R |
interleukin 4 receptor |
| chr6_+_112375275 | 0.96 |
ENST00000368666.2 ENST00000604763.1 ENST00000230529.5 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
| chr6_-_86352982 | 0.94 |
ENST00000369622.3 |
SYNCRIP |
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr3_-_55521323 | 0.94 |
ENST00000264634.4 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
| chr12_-_120687948 | 0.91 |
ENST00000458477.2 |
PXN |
paxillin |
| chr15_-_35088340 | 0.91 |
ENST00000290378.4 |
ACTC1 |
actin, alpha, cardiac muscle 1 |
| chr1_-_113249734 | 0.91 |
ENST00000484054.3 ENST00000369636.2 ENST00000369637.1 ENST00000285735.2 ENST00000369638.2 |
RHOC |
ras homolog family member C |
| chr11_-_89224488 | 0.85 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr7_-_19157248 | 0.84 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chr11_-_118436606 | 0.82 |
ENST00000530872.1 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr19_-_15560730 | 0.82 |
ENST00000389282.4 ENST00000263381.7 |
WIZ |
widely interspaced zinc finger motifs |
| chr1_-_113249948 | 0.78 |
ENST00000339083.7 ENST00000369642.3 |
RHOC |
ras homolog family member C |
| chr12_+_54447637 | 0.77 |
ENST00000609810.1 ENST00000430889.2 |
HOXC4 HOXC4 |
homeobox C4 Homeobox protein Hox-C4 |
| chr11_-_85779971 | 0.77 |
ENST00000393346.3 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr1_-_113249678 | 0.76 |
ENST00000369633.2 ENST00000425265.2 ENST00000369632.2 ENST00000436685.2 |
RHOC |
ras homolog family member C |
| chr9_-_13279589 | 0.75 |
ENST00000319217.7 |
MPDZ |
multiple PDZ domain protein |
| chr12_-_123756781 | 0.74 |
ENST00000544658.1 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chrX_-_148571884 | 0.74 |
ENST00000537071.1 |
IDS |
iduronate 2-sulfatase |
| chr1_+_157963063 | 0.73 |
ENST00000360089.4 ENST00000368173.3 ENST00000392272.2 |
KIRREL |
kin of IRRE like (Drosophila) |
| chr1_+_46668994 | 0.73 |
ENST00000371980.3 |
LURAP1 |
leucine rich adaptor protein 1 |
| chr5_+_151151471 | 0.72 |
ENST00000394123.3 ENST00000543466.1 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr12_-_56120865 | 0.70 |
ENST00000548898.1 ENST00000552067.1 |
CD63 |
CD63 molecule |
| chr12_-_6233828 | 0.70 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chr12_-_123756687 | 0.69 |
ENST00000261692.2 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
| chr6_+_151646800 | 0.67 |
ENST00000354675.6 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr11_-_85780086 | 0.67 |
ENST00000532317.1 ENST00000528256.1 ENST00000526033.1 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
| chr19_+_41305085 | 0.66 |
ENST00000303961.4 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr16_-_49890016 | 0.65 |
ENST00000563137.2 |
ZNF423 |
zinc finger protein 423 |
| chr19_+_34287751 | 0.63 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr12_-_56120838 | 0.60 |
ENST00000548160.1 |
CD63 |
CD63 molecule |
| chr20_-_36793663 | 0.60 |
ENST00000536701.1 ENST00000536724.1 |
TGM2 |
transglutaminase 2 |
| chr10_-_121356518 | 0.58 |
ENST00000369092.4 |
TIAL1 |
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr10_+_13628921 | 0.57 |
ENST00000378572.3 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr11_+_66406088 | 0.57 |
ENST00000310092.7 ENST00000396053.4 ENST00000408993.2 |
RBM4 |
RNA binding motif protein 4 |
| chr7_+_99686577 | 0.55 |
ENST00000303904.3 ENST00000419210.1 ENST00000418625.1 |
COPS6 |
COP9 signalosome subunit 6 |
| chr1_+_110091189 | 0.54 |
ENST00000369851.4 |
GNAI3 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr5_+_151151504 | 0.54 |
ENST00000356245.3 ENST00000507878.2 |
G3BP1 |
GTPase activating protein (SH3 domain) binding protein 1 |
| chr5_-_133512654 | 0.53 |
ENST00000522552.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr11_-_96239990 | 0.52 |
ENST00000511243.2 |
JRKL-AS1 |
JRKL antisense RNA 1 |
| chr5_-_133512683 | 0.52 |
ENST00000353411.6 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr10_+_13628933 | 0.52 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chr9_-_13279406 | 0.49 |
ENST00000546205.1 |
MPDZ |
multiple PDZ domain protein |
| chr6_-_136847610 | 0.49 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr16_+_1728305 | 0.48 |
ENST00000569765.1 |
HN1L |
hematological and neurological expressed 1-like |
| chr9_-_14722715 | 0.47 |
ENST00000380911.3 |
CER1 |
cerberus 1, DAN family BMP antagonist |
| chr19_-_3025614 | 0.45 |
ENST00000447365.2 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr3_+_184032919 | 0.44 |
ENST00000427845.1 ENST00000342981.4 ENST00000319274.6 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_23857698 | 0.44 |
ENST00000361729.2 |
E2F2 |
E2F transcription factor 2 |
| chrX_+_151867214 | 0.43 |
ENST00000329342.5 ENST00000412733.1 ENST00000457643.1 |
MAGEA6 |
melanoma antigen family A, 6 |
| chr3_+_8775466 | 0.43 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chr10_+_115438920 | 0.42 |
ENST00000429617.1 ENST00000369331.4 |
CASP7 |
caspase 7, apoptosis-related cysteine peptidase |
| chr17_-_4167142 | 0.42 |
ENST00000570535.1 ENST00000574367.1 ENST00000341657.4 ENST00000433651.1 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
| chr12_-_71314617 | 0.41 |
ENST00000283228.2 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr19_-_51471362 | 0.41 |
ENST00000376853.4 ENST00000424910.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr10_+_70091812 | 0.40 |
ENST00000265866.7 |
HNRNPH3 |
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr3_-_18466026 | 0.39 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr3_+_41240925 | 0.39 |
ENST00000396183.3 ENST00000349496.5 ENST00000453024.1 |
CTNNB1 |
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr12_+_62654155 | 0.39 |
ENST00000312635.6 ENST00000393654.3 ENST00000549237.1 |
USP15 |
ubiquitin specific peptidase 15 |
| chr18_+_33877654 | 0.39 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr12_-_57522813 | 0.39 |
ENST00000556155.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr19_+_41305406 | 0.38 |
ENST00000406058.2 ENST00000593726.1 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
| chr12_+_62654119 | 0.38 |
ENST00000353364.3 ENST00000549523.1 ENST00000280377.5 |
USP15 |
ubiquitin specific peptidase 15 |
| chr12_-_57940904 | 0.38 |
ENST00000550954.1 ENST00000434715.3 ENST00000546670.1 ENST00000543672.1 |
DCTN2 |
dynactin 2 (p50) |
| chrX_-_151903184 | 0.38 |
ENST00000357916.4 ENST00000393869.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chr19_-_47975417 | 0.38 |
ENST00000236877.6 |
SLC8A2 |
solute carrier family 8 (sodium/calcium exchanger), member 2 |
| chr3_+_41240986 | 0.38 |
ENST00000396185.3 |
CTNNB1 |
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr10_-_75415825 | 0.38 |
ENST00000394810.2 |
SYNPO2L |
synaptopodin 2-like |
| chr17_-_38978847 | 0.37 |
ENST00000269576.5 |
KRT10 |
keratin 10 |
| chr13_-_72441315 | 0.36 |
ENST00000305425.4 ENST00000313174.7 ENST00000354591.4 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chr12_-_57941004 | 0.36 |
ENST00000550750.1 ENST00000548249.1 |
DCTN2 |
dynactin 2 (p50) |
| chr10_+_70091847 | 0.35 |
ENST00000441000.2 ENST00000354695.5 |
HNRNPH3 |
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr1_+_21766641 | 0.35 |
ENST00000342104.5 |
NBPF3 |
neuroblastoma breakpoint family, member 3 |
| chr1_+_21766588 | 0.34 |
ENST00000454000.2 ENST00000318220.6 ENST00000318249.5 |
NBPF3 |
neuroblastoma breakpoint family, member 3 |
| chr19_-_51192661 | 0.34 |
ENST00000391813.1 |
SHANK1 |
SH3 and multiple ankyrin repeat domains 1 |
| chr3_+_192958914 | 0.34 |
ENST00000264735.2 ENST00000602513.1 |
HRASLS |
HRAS-like suppressor |
| chrX_+_148863584 | 0.34 |
ENST00000439010.2 ENST00000298974.5 ENST00000522429.1 ENST00000519822.1 |
MAGEA9 |
melanoma antigen family A, 9 |
| chrX_-_153881842 | 0.34 |
ENST00000369585.3 ENST00000247306.4 |
CTAG2 |
cancer/testis antigen 2 |
| chr7_-_102213030 | 0.34 |
ENST00000511313.1 ENST00000513438.1 ENST00000513506.1 |
POLR2J3 |
polymerase (RNA) II (DNA directed) polypeptide J3 |
| chr17_-_41132010 | 0.33 |
ENST00000409103.1 ENST00000360221.4 |
PTGES3L-AARSD1 |
PTGES3L-AARSD1 readthrough |
| chr13_-_108867846 | 0.33 |
ENST00000442234.1 |
LIG4 |
ligase IV, DNA, ATP-dependent |
| chr19_-_51471381 | 0.33 |
ENST00000594641.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr22_+_32750872 | 0.32 |
ENST00000397468.1 |
RFPL3 |
ret finger protein-like 3 |
| chr12_+_54402790 | 0.32 |
ENST00000040584.4 |
HOXC8 |
homeobox C8 |
| chr13_-_72440901 | 0.31 |
ENST00000359684.2 |
DACH1 |
dachshund homolog 1 (Drosophila) |
| chrX_-_151938171 | 0.31 |
ENST00000393902.3 ENST00000417212.1 ENST00000370278.3 |
MAGEA3 |
melanoma antigen family A, 3 |
| chr19_-_51336443 | 0.31 |
ENST00000598673.1 |
KLK15 |
kallikrein-related peptidase 15 |
| chr3_+_154797877 | 0.31 |
ENST00000462745.1 ENST00000493237.1 |
MME |
membrane metallo-endopeptidase |
| chr6_+_112375462 | 0.30 |
ENST00000361714.1 |
WISP3 |
WNT1 inducible signaling pathway protein 3 |
| chr11_+_71791693 | 0.29 |
ENST00000289488.2 ENST00000447974.1 |
LRTOMT |
leucine rich transmembrane and O-methyltransferase domain containing |
| chr16_+_21169976 | 0.29 |
ENST00000572258.1 ENST00000261388.3 ENST00000451578.2 ENST00000572599.1 ENST00000577162.1 |
TMEM159 |
transmembrane protein 159 |
| chrX_-_148669116 | 0.29 |
ENST00000243314.5 |
MAGEA9B |
melanoma antigen family A, 9B |
| chr11_-_414948 | 0.28 |
ENST00000530494.1 ENST00000528209.1 ENST00000431843.2 ENST00000528058.1 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr7_-_128045984 | 0.28 |
ENST00000470772.1 ENST00000480861.1 ENST00000496200.1 |
IMPDH1 |
IMP (inosine 5'-monophosphate) dehydrogenase 1 |
| chr17_-_39728303 | 0.28 |
ENST00000588431.1 ENST00000246662.4 |
KRT9 |
keratin 9 |
| chr16_+_1728257 | 0.28 |
ENST00000248098.3 ENST00000562684.1 ENST00000561516.1 ENST00000382711.5 ENST00000566742.1 |
HN1L |
hematological and neurological expressed 1-like |
| chr11_+_34073269 | 0.28 |
ENST00000389645.3 |
CAPRIN1 |
cell cycle associated protein 1 |
| chr1_-_226111929 | 0.28 |
ENST00000343818.6 ENST00000432920.2 |
PYCR2 RP4-559A3.7 |
pyrroline-5-carboxylate reductase family, member 2 Uncharacterized protein |
| chr2_+_48757278 | 0.27 |
ENST00000404752.1 ENST00000406226.1 |
STON1 |
stonin 1 |
| chr19_+_3366547 | 0.27 |
ENST00000341919.3 ENST00000590282.1 ENST00000443272.2 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr5_-_142065223 | 0.27 |
ENST00000378046.1 |
FGF1 |
fibroblast growth factor 1 (acidic) |
| chr3_+_154797428 | 0.27 |
ENST00000460393.1 |
MME |
membrane metallo-endopeptidase |
| chr2_+_106468204 | 0.27 |
ENST00000425756.1 ENST00000393349.2 |
NCK2 |
NCK adaptor protein 2 |
| chr1_-_201346761 | 0.27 |
ENST00000455702.1 ENST00000422165.1 ENST00000367318.5 ENST00000367320.2 ENST00000438742.1 ENST00000412633.1 ENST00000458432.2 ENST00000421663.2 ENST00000367322.1 ENST00000509001.1 |
TNNT2 |
troponin T type 2 (cardiac) |
| chr5_-_179227540 | 0.26 |
ENST00000520875.1 |
MGAT4B |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr18_-_3874247 | 0.26 |
ENST00000581699.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr12_+_26348246 | 0.25 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr12_+_26348429 | 0.25 |
ENST00000242729.2 |
SSPN |
sarcospan |
| chr12_-_16759711 | 0.25 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr19_+_44085189 | 0.24 |
ENST00000562365.2 |
PINLYP |
phospholipase A2 inhibitor and LY6/PLAUR domain containing |
| chr1_+_43148059 | 0.24 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr11_+_2323349 | 0.24 |
ENST00000381121.3 |
TSPAN32 |
tetraspanin 32 |
| chr16_-_4588469 | 0.24 |
ENST00000588381.1 ENST00000563332.2 |
CDIP1 |
cell death-inducing p53 target 1 |
| chrX_-_151903101 | 0.24 |
ENST00000393900.3 |
MAGEA12 |
melanoma antigen family A, 12 |
| chr5_+_150591678 | 0.23 |
ENST00000523466.1 |
GM2A |
GM2 ganglioside activator |
| chr11_-_35547151 | 0.23 |
ENST00000378878.3 ENST00000529303.1 ENST00000278360.3 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr16_-_31439735 | 0.23 |
ENST00000287490.4 |
COX6A2 |
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr2_-_230786619 | 0.22 |
ENST00000389045.3 ENST00000409677.1 |
TRIP12 |
thyroid hormone receptor interactor 12 |
| chr8_-_134115118 | 0.22 |
ENST00000395352.3 ENST00000338087.5 |
SLA |
Src-like-adaptor |
| chrX_-_151307020 | 0.21 |
ENST00000370323.4 ENST00000244096.3 ENST00000427322.2 |
MAGEA10 |
melanoma antigen family A, 10 |
| chr12_+_132413739 | 0.21 |
ENST00000443358.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr1_+_154975110 | 0.21 |
ENST00000535420.1 ENST00000368426.3 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
| chr3_-_38992052 | 0.20 |
ENST00000302328.3 ENST00000450244.1 ENST00000444237.2 |
SCN11A |
sodium channel, voltage-gated, type XI, alpha subunit |
| chr5_+_131396222 | 0.20 |
ENST00000296870.2 |
IL3 |
interleukin 3 (colony-stimulating factor, multiple) |
| chr7_-_14028488 | 0.20 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr12_+_132413765 | 0.20 |
ENST00000376649.3 ENST00000322060.5 |
PUS1 |
pseudouridylate synthase 1 |
| chr16_+_28889801 | 0.20 |
ENST00000395503.4 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chrX_-_153847522 | 0.20 |
ENST00000328435.2 ENST00000359887.4 |
CTAG1B |
cancer/testis antigen 1B |
| chr11_+_17756279 | 0.20 |
ENST00000265969.6 |
KCNC1 |
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
| chrX_+_153813407 | 0.20 |
ENST00000443287.2 ENST00000333128.3 |
CTAG1A |
cancer/testis antigen 1A |
| chr16_+_28889703 | 0.19 |
ENST00000357084.3 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
| chr5_-_35195338 | 0.19 |
ENST00000509839.1 |
PRLR |
prolactin receptor |
| chr2_-_54087066 | 0.19 |
ENST00000394705.2 ENST00000352846.3 ENST00000406625.2 |
GPR75 GPR75-ASB3 ASB3 |
G protein-coupled receptor 75 GPR75-ASB3 readthrough Ankyrin repeat and SOCS box protein 3 |
| chr19_-_3029011 | 0.19 |
ENST00000590536.1 ENST00000587137.1 ENST00000455444.2 ENST00000262953.6 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr12_+_16064258 | 0.19 |
ENST00000524480.1 ENST00000531803.1 ENST00000532964.1 |
DERA |
deoxyribose-phosphate aldolase (putative) |
| chr1_-_21377383 | 0.18 |
ENST00000374935.3 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr1_-_1711508 | 0.18 |
ENST00000378625.1 |
NADK |
NAD kinase |
| chr11_+_20044600 | 0.17 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chrX_-_151922340 | 0.17 |
ENST00000370284.1 ENST00000543232.1 ENST00000393876.1 ENST00000393872.3 |
MAGEA2 |
melanoma antigen family A, 2 |
| chr18_-_3874752 | 0.17 |
ENST00000534970.1 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr1_-_204380919 | 0.17 |
ENST00000367188.4 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
| chr11_-_129872712 | 0.17 |
ENST00000358825.5 ENST00000360871.3 ENST00000528746.1 |
PRDM10 |
PR domain containing 10 |
| chr11_+_125034586 | 0.17 |
ENST00000298282.9 |
PKNOX2 |
PBX/knotted 1 homeobox 2 |
| chr8_+_32406137 | 0.17 |
ENST00000521670.1 |
NRG1 |
neuregulin 1 |
| chr18_-_3874271 | 0.16 |
ENST00000400149.3 ENST00000400155.1 ENST00000400150.3 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_+_147560042 | 0.16 |
ENST00000281321.3 |
POU4F2 |
POU class 4 homeobox 2 |
| chr1_-_94586651 | 0.15 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr14_+_105155925 | 0.15 |
ENST00000330634.7 ENST00000398337.4 ENST00000392634.4 |
INF2 |
inverted formin, FH2 and WH2 domain containing |
| chr7_-_38398721 | 0.15 |
ENST00000390346.2 |
TRGV3 |
T cell receptor gamma variable 3 |
| chr11_+_34073195 | 0.14 |
ENST00000341394.4 |
CAPRIN1 |
cell cycle associated protein 1 |
| chrX_+_128674213 | 0.14 |
ENST00000371113.4 ENST00000357121.5 |
OCRL |
oculocerebrorenal syndrome of Lowe |
| chr19_-_9006766 | 0.14 |
ENST00000599436.1 |
MUC16 |
mucin 16, cell surface associated |
| chr12_-_49351148 | 0.13 |
ENST00000398092.4 ENST00000539611.1 |
RP11-302B13.5 ARF3 |
ADP-ribosylation factor 3 ADP-ribosylation factor 3 |
| chr2_+_105471969 | 0.13 |
ENST00000361360.2 |
POU3F3 |
POU class 3 homeobox 3 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.2 | 2.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 4.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 4.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 3.6 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 5.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.1 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 3.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.9 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.7 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 2.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.7 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.7 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 2.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 4.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 1.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 1.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 3.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 4.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 3.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.2 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 1.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 4.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 2.5 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.1 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 1.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.4 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 2.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 9.9 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.4 | 1.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.3 | 4.5 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.3 | 3.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.3 | 1.2 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.2 | 1.4 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.2 | 3.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.2 | 2.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.2 | 0.9 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 1.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 1.2 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.1 | 2.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) Flemming body(GO:0090543) |
| 0.1 | 0.4 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 4.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.1 | 1.0 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.1 | 2.2 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.1 | 1.3 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 1.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 2.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 8.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 5.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 1.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 0.3 | GO:0032807 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 3.6 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 2.4 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.8 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.6 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.2 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 3.3 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 2.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.8 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.4 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.8 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.9 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 1.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 9.9 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.6 | 4.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.6 | 1.7 | GO:0042700 | luteinizing hormone signaling pathway(GO:0042700) |
| 0.5 | 3.6 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.5 | 4.5 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.5 | 0.9 | GO:0003213 | cardiac right atrium morphogenesis(GO:0003213) |
| 0.4 | 1.8 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.4 | 0.8 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.4 | 2.5 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.4 | 1.6 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.4 | 2.7 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.4 | 2.6 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.4 | 1.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.4 | 3.9 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.3 | 2.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 0.8 | GO:0061316 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in heart development(GO:0061316) regulation of centromeric sister chromatid cohesion(GO:0070602) renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) positive regulation of determination of dorsal identity(GO:2000017) |
| 0.2 | 2.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.2 | 1.4 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.2 | 0.6 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.2 | 1.0 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) response to odorant(GO:1990834) |
| 0.2 | 2.1 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) |
| 0.2 | 0.8 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 1.2 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.2 | 1.0 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.2 | 0.3 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.5 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.2 | 0.9 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.4 | GO:0050894 | determination of affect(GO:0050894) |
| 0.1 | 0.4 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.1 | 3.6 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.4 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 1.2 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.1 | 0.4 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 1.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.4 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.1 | 3.3 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.1 | 2.2 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.1 | 0.3 | GO:0051102 | DNA ligation involved in DNA recombination(GO:0051102) |
| 0.1 | 1.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.4 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 1.0 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.3 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 1.4 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 | 0.4 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.8 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.1 | 0.7 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.1 | 1.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.2 | GO:0061074 | regulation of neural retina development(GO:0061074) regulation of retina development in camera-type eye(GO:1902866) |
| 0.1 | 5.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.4 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.1 | 0.4 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.3 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.6 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 1.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.1 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.4 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:2000507 | cap-dependent translational initiation(GO:0002191) positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.3 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.2 | GO:1903912 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.0 | 0.3 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.7 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.2 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 3.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.1 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.1 | GO:0072240 | thick ascending limb development(GO:0072023) DCT cell differentiation(GO:0072069) metanephric thick ascending limb development(GO:0072233) metanephric DCT cell differentiation(GO:0072240) |
| 0.0 | 0.7 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.0 | 0.9 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.3 | GO:0060117 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.0 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.8 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 1.5 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 3.3 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.6 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.1 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.9 | 2.6 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.8 | 2.4 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.7 | 3.9 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.6 | 4.5 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.5 | 1.6 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.4 | 4.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.4 | 1.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.3 | 1.0 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) |
| 0.3 | 0.8 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.3 | 2.4 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.2 | 3.6 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 3.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.9 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 0.6 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.2 | 1.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.4 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.1 | 0.4 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.1 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.8 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 1.4 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 5.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 1.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 1.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.7 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.0 | 1.0 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.5 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 3.4 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 1.4 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 12.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0031691 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 1.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.9 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 1.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.6 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.8 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.8 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.4 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 1.4 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 2.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |