Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
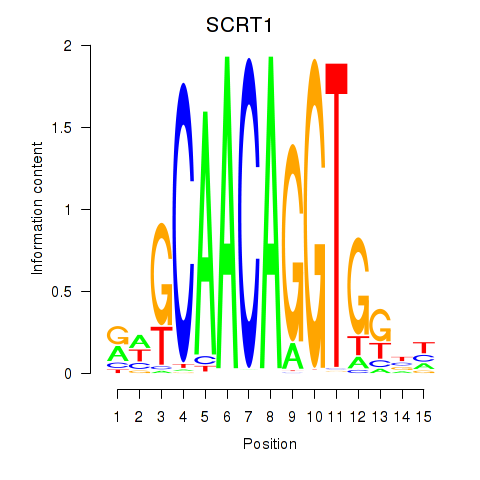
Results for SCRT1_SCRT2
Z-value: 1.39
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | SCRT1 |
|
SCRT2
|
ENSG00000215397.3 | SCRT2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SCRT1 | hg19_v2_chr8_-_145559943_145559943 | -0.03 | 9.2e-01 | Click! |
Activity profile of SCRT1_SCRT2 motif
Sorted Z-values of SCRT1_SCRT2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SCRT1_SCRT2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_41258786 | 6.11 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr6_+_121756809 | 5.26 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr6_-_31697255 | 4.10 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr1_-_95392635 | 3.66 |
ENST00000538964.1 ENST00000394202.4 ENST00000370206.4 |
CNN3 |
calponin 3, acidic |
| chr2_-_175870085 | 3.61 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr6_-_31697563 | 3.41 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr5_+_15500280 | 2.64 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr1_-_173176452 | 2.48 |
ENST00000281834.3 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chrX_+_51636629 | 2.21 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chrX_+_51927919 | 2.00 |
ENST00000416960.1 |
MAGED4 |
melanoma antigen family D, 4 |
| chrX_-_51812268 | 1.99 |
ENST00000486010.1 ENST00000497164.1 ENST00000360134.6 ENST00000485287.1 ENST00000335504.5 ENST00000431659.1 |
MAGED4B |
melanoma antigen family D, 4B |
| chr7_+_130126165 | 1.98 |
ENST00000427521.1 ENST00000416162.2 ENST00000378576.4 |
MEST |
mesoderm specific transcript |
| chr7_+_130126012 | 1.91 |
ENST00000341441.5 |
MEST |
mesoderm specific transcript |
| chr15_-_30113676 | 1.73 |
ENST00000400011.2 |
TJP1 |
tight junction protein 1 |
| chrX_+_51928002 | 1.72 |
ENST00000375626.3 |
MAGED4 |
melanoma antigen family D, 4 |
| chr11_+_32112431 | 1.63 |
ENST00000054950.3 |
RCN1 |
reticulocalbin 1, EF-hand calcium binding domain |
| chr4_-_119757239 | 1.59 |
ENST00000280551.6 |
SEC24D |
SEC24 family member D |
| chr5_-_111091948 | 1.52 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr11_+_69455855 | 1.47 |
ENST00000227507.2 ENST00000536559.1 |
CCND1 |
cyclin D1 |
| chr15_-_30114231 | 1.46 |
ENST00000356107.6 ENST00000545208.2 |
TJP1 |
tight junction protein 1 |
| chr4_-_149363662 | 1.46 |
ENST00000355292.3 ENST00000358102.3 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chr3_-_114343768 | 1.38 |
ENST00000393785.2 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr4_-_52904425 | 1.35 |
ENST00000535450.1 |
SGCB |
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr12_-_13153139 | 1.34 |
ENST00000536942.1 ENST00000014930.4 |
HEBP1 |
heme binding protein 1 |
| chr7_-_122526499 | 1.23 |
ENST00000412584.2 |
CADPS2 |
Ca++-dependent secretion activator 2 |
| chr7_-_122526799 | 1.17 |
ENST00000334010.7 ENST00000313070.7 |
CADPS2 |
Ca++-dependent secretion activator 2 |
| chr1_-_201391149 | 1.17 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr19_-_11373128 | 1.17 |
ENST00000294618.7 |
DOCK6 |
dedicator of cytokinesis 6 |
| chr9_+_125703282 | 1.12 |
ENST00000373647.4 ENST00000402311.1 |
RABGAP1 |
RAB GTPase activating protein 1 |
| chr11_+_842928 | 1.07 |
ENST00000397408.1 |
TSPAN4 |
tetraspanin 4 |
| chr19_-_58609570 | 1.07 |
ENST00000600845.1 ENST00000240727.6 ENST00000600897.1 ENST00000421612.2 ENST00000601063.1 ENST00000601144.1 |
ZSCAN18 |
zinc finger and SCAN domain containing 18 |
| chr4_-_119757322 | 1.07 |
ENST00000379735.5 |
SEC24D |
SEC24 family member D |
| chr1_-_94312706 | 1.02 |
ENST00000370244.1 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr1_+_150245099 | 0.99 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr8_+_98881268 | 0.97 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr16_+_30383613 | 0.96 |
ENST00000568749.1 |
MYLPF |
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr4_-_10118348 | 0.95 |
ENST00000502702.1 |
WDR1 |
WD repeat domain 1 |
| chr20_-_43977055 | 0.92 |
ENST00000372733.3 ENST00000537976.1 |
SDC4 |
syndecan 4 |
| chr12_-_71003568 | 0.86 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr11_+_842808 | 0.85 |
ENST00000397397.2 ENST00000397411.2 ENST00000397396.1 |
TSPAN4 |
tetraspanin 4 |
| chr5_+_72921983 | 0.84 |
ENST00000296794.6 ENST00000545377.1 ENST00000513042.2 ENST00000287898.5 ENST00000509848.1 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr16_-_53537105 | 0.84 |
ENST00000568596.1 ENST00000570004.1 ENST00000564497.1 ENST00000300245.4 ENST00000394657.7 |
AKTIP |
AKT interacting protein |
| chr1_-_52456352 | 0.82 |
ENST00000371655.3 |
RAB3B |
RAB3B, member RAS oncogene family |
| chr22_-_22090064 | 0.81 |
ENST00000339468.3 |
YPEL1 |
yippee-like 1 (Drosophila) |
| chr11_-_111794446 | 0.79 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_109289279 | 0.76 |
ENST00000370008.3 |
STXBP3 |
syntaxin binding protein 3 |
| chr11_-_61659006 | 0.72 |
ENST00000278829.2 |
FADS3 |
fatty acid desaturase 3 |
| chrX_+_15518923 | 0.68 |
ENST00000348343.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr10_-_101945771 | 0.67 |
ENST00000370408.2 ENST00000407654.3 |
ERLIN1 |
ER lipid raft associated 1 |
| chr5_+_125758813 | 0.65 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr7_+_99699280 | 0.64 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr6_-_83903600 | 0.63 |
ENST00000506587.1 ENST00000507554.1 |
PGM3 |
phosphoglucomutase 3 |
| chr5_+_125758865 | 0.62 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr11_+_65779283 | 0.58 |
ENST00000312134.2 |
CST6 |
cystatin E/M |
| chr11_+_6624955 | 0.56 |
ENST00000299421.4 ENST00000537806.1 |
ILK |
integrin-linked kinase |
| chr2_-_24583583 | 0.56 |
ENST00000355123.4 |
ITSN2 |
intersectin 2 |
| chr7_+_99699179 | 0.56 |
ENST00000438383.1 ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr21_-_35987438 | 0.54 |
ENST00000313806.4 |
RCAN1 |
regulator of calcineurin 1 |
| chr1_+_110162448 | 0.51 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr4_+_41540160 | 0.51 |
ENST00000503057.1 ENST00000511496.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr11_+_6624970 | 0.50 |
ENST00000420936.2 ENST00000528995.1 |
ILK |
integrin-linked kinase |
| chr13_+_58206655 | 0.49 |
ENST00000377918.3 |
PCDH17 |
protocadherin 17 |
| chr2_-_178753465 | 0.48 |
ENST00000389683.3 |
PDE11A |
phosphodiesterase 11A |
| chr17_-_33760164 | 0.46 |
ENST00000445092.1 ENST00000394562.1 ENST00000447040.2 |
SLFN12 |
schlafen family member 12 |
| chr5_-_73936451 | 0.44 |
ENST00000537006.1 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
| chr14_+_67708137 | 0.44 |
ENST00000556345.1 ENST00000555925.1 ENST00000557783.1 |
MPP5 |
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr14_+_102829300 | 0.44 |
ENST00000359520.7 |
TECPR2 |
tectonin beta-propeller repeat containing 2 |
| chr11_-_117747607 | 0.43 |
ENST00000540359.1 ENST00000539526.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr11_-_117748138 | 0.42 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr17_-_33760269 | 0.41 |
ENST00000452764.3 |
SLFN12 |
schlafen family member 12 |
| chr5_+_32710736 | 0.41 |
ENST00000415685.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr4_-_141348789 | 0.40 |
ENST00000414773.1 |
CLGN |
calmegin |
| chr8_-_116681221 | 0.40 |
ENST00000395715.3 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chrX_+_55744228 | 0.39 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr19_+_40877583 | 0.39 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chr20_+_57427765 | 0.38 |
ENST00000371100.4 |
GNAS |
GNAS complex locus |
| chr7_+_12727250 | 0.38 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr9_+_114423615 | 0.37 |
ENST00000374293.4 |
GNG10 |
guanine nucleotide binding protein (G protein), gamma 10 |
| chr4_-_5890145 | 0.37 |
ENST00000397890.2 |
CRMP1 |
collapsin response mediator protein 1 |
| chr9_+_99212403 | 0.36 |
ENST00000375251.3 ENST00000375249.4 |
HABP4 |
hyaluronan binding protein 4 |
| chr5_+_138089100 | 0.36 |
ENST00000520339.1 ENST00000355078.5 ENST00000302763.7 ENST00000518910.1 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr4_-_10117949 | 0.35 |
ENST00000508079.1 |
WDR1 |
WD repeat domain 1 |
| chr11_+_61583721 | 0.34 |
ENST00000257261.6 |
FADS2 |
fatty acid desaturase 2 |
| chr3_-_146187088 | 0.34 |
ENST00000497985.1 |
PLSCR2 |
phospholipid scramblase 2 |
| chr3_-_79816965 | 0.33 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr3_+_10857885 | 0.33 |
ENST00000254488.2 ENST00000454147.1 |
SLC6A11 |
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr14_+_67707826 | 0.33 |
ENST00000261681.4 |
MPP5 |
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr22_-_22090043 | 0.33 |
ENST00000403503.1 |
YPEL1 |
yippee-like 1 (Drosophila) |
| chr19_-_46974741 | 0.32 |
ENST00000313683.10 ENST00000602246.1 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr2_-_24583314 | 0.31 |
ENST00000443927.1 ENST00000406921.3 ENST00000412011.1 |
ITSN2 |
intersectin 2 |
| chr19_-_46974664 | 0.30 |
ENST00000438932.2 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
| chr5_-_59064458 | 0.30 |
ENST00000502575.1 ENST00000507116.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr5_+_74011328 | 0.30 |
ENST00000513336.1 |
HEXB |
hexosaminidase B (beta polypeptide) |
| chr16_-_30022293 | 0.29 |
ENST00000565273.1 ENST00000567332.2 ENST00000350119.4 |
DOC2A |
double C2-like domains, alpha |
| chr14_+_23775971 | 0.28 |
ENST00000250405.5 |
BCL2L2 |
BCL2-like 2 |
| chr3_+_133465228 | 0.25 |
ENST00000482271.1 ENST00000264998.3 |
TF |
transferrin |
| chr8_+_22429205 | 0.25 |
ENST00000520207.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr1_-_154928562 | 0.25 |
ENST00000368463.3 ENST00000539880.1 ENST00000542459.1 ENST00000368460.3 ENST00000368465.1 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
| chr3_+_141144954 | 0.24 |
ENST00000441582.2 ENST00000321464.5 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr11_+_27062272 | 0.24 |
ENST00000529202.1 ENST00000533566.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_-_212588157 | 0.24 |
ENST00000261455.4 ENST00000535273.1 |
TMEM206 |
transmembrane protein 206 |
| chr9_-_116840728 | 0.22 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr6_+_28092338 | 0.22 |
ENST00000340487.4 |
ZSCAN16 |
zinc finger and SCAN domain containing 16 |
| chr17_-_79805146 | 0.21 |
ENST00000415593.1 |
P4HB |
prolyl 4-hydroxylase, beta polypeptide |
| chr12_+_81471816 | 0.20 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr6_+_71122974 | 0.20 |
ENST00000418814.2 |
FAM135A |
family with sequence similarity 135, member A |
| chr2_-_224467093 | 0.19 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr7_+_114562172 | 0.19 |
ENST00000393486.1 ENST00000257724.3 |
MDFIC |
MyoD family inhibitor domain containing |
| chr17_-_32484313 | 0.18 |
ENST00000359872.6 |
ASIC2 |
acid-sensing (proton-gated) ion channel 2 |
| chr19_-_5838768 | 0.17 |
ENST00000527106.1 ENST00000531199.1 ENST00000529165.1 |
FUT6 |
fucosyltransferase 6 (alpha (1,3) fucosyltransferase) |
| chr4_+_980785 | 0.17 |
ENST00000247933.4 ENST00000453894.1 |
IDUA |
iduronidase, alpha-L- |
| chr1_+_44115814 | 0.16 |
ENST00000372396.3 |
KDM4A |
lysine (K)-specific demethylase 4A |
| chr8_+_132952112 | 0.16 |
ENST00000520362.1 ENST00000519656.1 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
| chrX_+_118370211 | 0.16 |
ENST00000217971.7 |
PGRMC1 |
progesterone receptor membrane component 1 |
| chr3_-_186080012 | 0.15 |
ENST00000544847.1 ENST00000265022.3 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
| chr11_-_117747434 | 0.15 |
ENST00000529335.2 ENST00000530956.1 ENST00000260282.4 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr12_-_57824739 | 0.14 |
ENST00000347140.3 ENST00000402412.1 |
R3HDM2 |
R3H domain containing 2 |
| chr4_-_120133661 | 0.14 |
ENST00000503243.1 ENST00000326780.3 |
RP11-455G16.1 |
Uncharacterized protein |
| chr9_+_100174344 | 0.13 |
ENST00000422139.2 |
TDRD7 |
tudor domain containing 7 |
| chr1_-_156918806 | 0.13 |
ENST00000315174.8 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr6_+_108977520 | 0.13 |
ENST00000540898.1 |
FOXO3 |
forkhead box O3 |
| chr16_+_25123041 | 0.12 |
ENST00000399069.3 ENST00000380966.4 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
| chr6_-_31080336 | 0.12 |
ENST00000259870.3 |
C6orf15 |
chromosome 6 open reading frame 15 |
| chr22_+_40742512 | 0.12 |
ENST00000454266.2 ENST00000342312.6 |
ADSL |
adenylosuccinate lyase |
| chr17_-_2415169 | 0.11 |
ENST00000263092.6 ENST00000538844.1 ENST00000576976.1 |
METTL16 |
methyltransferase like 16 |
| chr19_-_18391708 | 0.11 |
ENST00000600972.1 |
JUND |
jun D proto-oncogene |
| chr17_-_40288449 | 0.11 |
ENST00000552162.1 ENST00000550504.1 |
RAB5C |
RAB5C, member RAS oncogene family |
| chr3_-_62861012 | 0.11 |
ENST00000357948.3 ENST00000383710.4 |
CADPS |
Ca++-dependent secretion activator |
| chr15_+_84904525 | 0.10 |
ENST00000510439.2 |
GOLGA6L4 |
golgin A6 family-like 4 |
| chr2_-_167232484 | 0.10 |
ENST00000375387.4 ENST00000303354.6 ENST00000409672.1 |
SCN9A |
sodium channel, voltage-gated, type IX, alpha subunit |
| chr16_-_25122785 | 0.10 |
ENST00000563962.1 ENST00000569920.1 |
RP11-449H11.1 |
RP11-449H11.1 |
| chr17_+_63133587 | 0.10 |
ENST00000449996.3 ENST00000262406.9 |
RGS9 |
regulator of G-protein signaling 9 |
| chr10_-_95209 | 0.10 |
ENST00000332708.5 ENST00000309812.4 |
TUBB8 |
tubulin, beta 8 class VIII |
| chr1_-_224033596 | 0.09 |
ENST00000391878.2 ENST00000343537.7 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
| chr1_+_225965518 | 0.08 |
ENST00000304786.7 ENST00000366839.4 ENST00000366838.1 |
SRP9 |
signal recognition particle 9kDa |
| chr15_+_80351910 | 0.08 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr1_-_222885770 | 0.08 |
ENST00000355727.2 ENST00000340020.6 |
AIDA |
axin interactor, dorsalization associated |
| chr12_+_103981044 | 0.07 |
ENST00000388887.2 |
STAB2 |
stabilin 2 |
| chr1_+_43148625 | 0.07 |
ENST00000436427.1 |
YBX1 |
Y box binding protein 1 |
| chr7_+_94536898 | 0.07 |
ENST00000433360.1 ENST00000340694.4 ENST00000424654.1 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
| chr2_+_173600671 | 0.06 |
ENST00000409036.1 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr12_-_122907091 | 0.06 |
ENST00000358808.2 ENST00000361654.4 ENST00000539080.1 ENST00000537178.1 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
| chr21_-_34852304 | 0.06 |
ENST00000542230.2 |
TMEM50B |
transmembrane protein 50B |
| chr10_-_47222824 | 0.05 |
ENST00000355232.3 |
AGAP10 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 10 |
| chr8_+_145438870 | 0.05 |
ENST00000527931.1 |
FAM203B |
family with sequence similarity 203, member B |
| chr4_+_87856191 | 0.04 |
ENST00000503477.1 |
AFF1 |
AF4/FMR2 family, member 1 |
| chr8_-_101718991 | 0.03 |
ENST00000517990.1 |
PABPC1 |
poly(A) binding protein, cytoplasmic 1 |
| chr19_+_30097181 | 0.03 |
ENST00000586420.1 ENST00000221770.3 ENST00000392279.3 ENST00000590688.1 |
POP4 |
processing of precursor 4, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr12_-_117318788 | 0.03 |
ENST00000550505.1 |
HRK |
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr6_-_34113856 | 0.02 |
ENST00000538487.2 |
GRM4 |
glutamate receptor, metabotropic 4 |
| chr1_-_204135450 | 0.02 |
ENST00000272190.8 ENST00000367195.2 |
REN |
renin |
| chr19_+_52264104 | 0.01 |
ENST00000340023.6 |
FPR2 |
formyl peptide receptor 2 |
| chr5_+_173763250 | 0.01 |
ENST00000515513.1 ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1 |
RP11-267A15.1 |
| chr6_+_71123107 | 0.01 |
ENST00000370479.3 ENST00000505769.1 ENST00000515323.1 ENST00000515280.1 ENST00000507085.1 ENST00000457062.2 ENST00000361499.3 |
FAM135A |
family with sequence similarity 135, member A |
| chr22_-_19466732 | 0.01 |
ENST00000263202.10 ENST00000360834.4 |
UFD1L |
ubiquitin fusion degradation 1 like (yeast) |
| chr19_+_12862604 | 0.00 |
ENST00000553030.1 |
BEST2 |
bestrophin 2 |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 4.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 5.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 4.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 2.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.8 | 5.3 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 1.1 | 3.2 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.8 | 2.5 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.5 | 2.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.5 | 7.5 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.3 | 1.3 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.3 | 2.6 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 1.1 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.2 | 0.9 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.2 | 0.8 | GO:0051582 | peptidyl-cysteine methylation(GO:0018125) positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.2 | 3.7 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 1.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.7 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.1 | 0.6 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.4 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:0021836 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 3.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 1.3 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.1 | 2.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 2.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.8 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.1 | 0.5 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.1 | 1.2 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.7 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.1 | 0.3 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.1 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.1 | 0.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 1.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 1.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.0 | 0.6 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.3 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.5 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.2 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 3.9 | GO:0007498 | mesoderm development(GO:0007498) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.3 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.2 | GO:1900113 | histone H3-K36 demethylation(GO:0070544) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 0.0 | 0.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 1.0 | GO:0031102 | neuron projection regeneration(GO:0031102) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.0 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 1.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.5 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.2 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.7 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) |
| 0.0 | 1.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.2 | 2.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.7 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.5 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.4 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 3.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 1.5 | 7.5 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 1.1 | 5.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.1 | 0.3 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 2.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.9 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 3.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.5 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 1.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.7 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 1.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.8 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 2.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 1.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 2.2 | GO:0005496 | steroid binding(GO:0005496) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 3.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.3 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.3 | 1.3 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.2 | 0.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 5.9 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 3.2 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.2 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.4 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 0.8 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.1 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 2.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 1.0 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 1.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.2 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 7.8 | GO:0005815 | microtubule organizing center(GO:0005815) |