Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
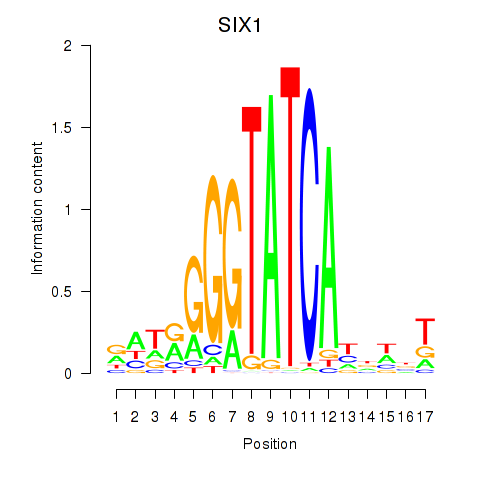
Results for SIX1_SIX3_SIX2
Z-value: 0.89
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX1 |
|
SIX3
|
ENSG00000138083.3 | SIX3 |
|
SIX2
|
ENSG00000170577.7 | SIX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX3 | hg19_v2_chr2_+_45168875_45168916 | 0.50 | 5.0e-02 | Click! |
| SIX1 | hg19_v2_chr14_-_61116168_61116180 | 0.17 | 5.3e-01 | Click! |
| SIX2 | hg19_v2_chr2_-_45236540_45236577 | 0.05 | 8.4e-01 | Click! |
Activity profile of SIX1_SIX3_SIX2 motif
Sorted Z-values of SIX1_SIX3_SIX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX1_SIX3_SIX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_46679144 | 2.32 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr13_-_46679185 | 2.31 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr16_-_28550348 | 1.57 |
ENST00000324873.6 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr5_+_125758865 | 1.54 |
ENST00000542322.1 ENST00000544396.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr16_-_28550320 | 1.45 |
ENST00000395641.2 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
| chr5_+_125758813 | 1.44 |
ENST00000285689.3 ENST00000515200.1 |
GRAMD3 |
GRAM domain containing 3 |
| chr3_-_149095652 | 1.32 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr10_+_48255253 | 1.22 |
ENST00000357718.4 ENST00000344416.5 ENST00000456111.2 ENST00000374258.3 |
ANXA8 AL591684.1 |
annexin A8 Protein LOC100996760 |
| chr11_-_118134997 | 1.12 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr10_-_47173994 | 1.09 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr10_+_47746929 | 1.08 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr6_-_138428613 | 1.07 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chr1_+_15256230 | 1.05 |
ENST00000376028.4 ENST00000400798.2 |
KAZN |
kazrin, periplakin interacting protein |
| chr12_-_52845910 | 1.02 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr18_+_55816546 | 0.96 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr3_-_111314230 | 0.94 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr10_+_5005445 | 0.90 |
ENST00000380872.4 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr2_-_113594279 | 0.73 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr2_+_211421262 | 0.71 |
ENST00000233072.5 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chr6_-_136847099 | 0.69 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr10_+_7745232 | 0.66 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr10_+_7745303 | 0.65 |
ENST00000429820.1 ENST00000379587.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr12_+_81471816 | 0.64 |
ENST00000261206.3 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
| chr14_+_95047725 | 0.61 |
ENST00000554760.1 ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr17_+_4675175 | 0.60 |
ENST00000270560.3 |
TM4SF5 |
transmembrane 4 L six family member 5 |
| chr11_+_70244510 | 0.59 |
ENST00000346329.3 ENST00000301843.8 ENST00000376561.3 |
CTTN |
cortactin |
| chr16_+_56642041 | 0.58 |
ENST00000245185.5 |
MT2A |
metallothionein 2A |
| chr8_-_48651648 | 0.57 |
ENST00000408965.3 |
CEBPD |
CCAAT/enhancer binding protein (C/EBP), delta |
| chr10_+_5005598 | 0.56 |
ENST00000442997.1 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr3_+_100211412 | 0.53 |
ENST00000323523.4 ENST00000403410.1 ENST00000449609.1 |
TMEM45A |
transmembrane protein 45A |
| chr6_+_28048753 | 0.50 |
ENST00000377325.1 |
ZNF165 |
zinc finger protein 165 |
| chr20_+_4667094 | 0.49 |
ENST00000424424.1 ENST00000457586.1 |
PRNP |
prion protein |
| chr11_-_112034831 | 0.48 |
ENST00000280357.7 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
| chr12_+_19358228 | 0.46 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr3_-_11685345 | 0.45 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chrX_+_152907913 | 0.44 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chr2_+_211342432 | 0.44 |
ENST00000430249.2 |
CPS1 |
carbamoyl-phosphate synthase 1, mitochondrial |
| chr19_-_48389651 | 0.44 |
ENST00000222002.3 |
SULT2A1 |
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr1_+_207277590 | 0.43 |
ENST00000367070.3 |
C4BPA |
complement component 4 binding protein, alpha |
| chr4_+_155484155 | 0.40 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr8_-_18666360 | 0.39 |
ENST00000286485.8 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
| chr2_+_234826016 | 0.38 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr3_+_111718036 | 0.35 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr12_+_57623869 | 0.35 |
ENST00000414700.3 ENST00000557703.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr14_-_94759361 | 0.35 |
ENST00000393096.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr17_+_1665345 | 0.33 |
ENST00000576406.1 ENST00000571149.1 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr19_+_2867325 | 0.33 |
ENST00000307635.2 ENST00000586426.1 |
ZNF556 |
zinc finger protein 556 |
| chr6_+_131894284 | 0.33 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr1_+_73771844 | 0.32 |
ENST00000440762.1 ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1 |
RP4-598G3.1 |
| chr6_-_136847610 | 0.32 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr3_+_111717600 | 0.32 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr4_-_156298028 | 0.29 |
ENST00000433024.1 ENST00000379248.2 |
MAP9 |
microtubule-associated protein 9 |
| chr1_+_145293371 | 0.29 |
ENST00000342960.5 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr7_+_29519486 | 0.29 |
ENST00000409041.4 |
CHN2 |
chimerin 2 |
| chr16_+_28763108 | 0.29 |
ENST00000357796.3 ENST00000550983.1 |
NPIPB9 |
nuclear pore complex interacting protein family, member B9 |
| chr2_-_114514181 | 0.27 |
ENST00000409342.1 |
SLC35F5 |
solute carrier family 35, member F5 |
| chr7_+_139025875 | 0.27 |
ENST00000297534.6 |
C7orf55 |
chromosome 7 open reading frame 55 |
| chr20_+_34700333 | 0.27 |
ENST00000441639.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr4_-_156297949 | 0.27 |
ENST00000515654.1 |
MAP9 |
microtubule-associated protein 9 |
| chr11_-_62521614 | 0.26 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr3_+_42850959 | 0.25 |
ENST00000442925.1 ENST00000422265.1 ENST00000497921.1 ENST00000426937.1 |
ACKR2 KRBOX1 |
atypical chemokine receptor 2 KRAB box domain containing 1 |
| chr3_+_111718173 | 0.25 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr4_-_156298087 | 0.25 |
ENST00000311277.4 |
MAP9 |
microtubule-associated protein 9 |
| chr19_+_45445491 | 0.24 |
ENST00000592954.1 ENST00000419266.2 ENST00000589057.1 |
APOC4 APOC4-APOC2 |
apolipoprotein C-IV APOC4-APOC2 readthrough (NMD candidate) |
| chr14_-_94759595 | 0.23 |
ENST00000261994.4 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr12_+_57624085 | 0.23 |
ENST00000553474.1 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_+_57624119 | 0.23 |
ENST00000555773.1 ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2 |
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr19_+_36605850 | 0.22 |
ENST00000221855.3 |
TBCB |
tubulin folding cofactor B |
| chr1_+_93913665 | 0.22 |
ENST00000271234.7 ENST00000370256.4 ENST00000260506.8 |
FNBP1L |
formin binding protein 1-like |
| chr11_+_61520075 | 0.22 |
ENST00000278836.5 |
MYRF |
myelin regulatory factor |
| chr10_+_5238793 | 0.22 |
ENST00000263126.1 |
AKR1C4 |
aldo-keto reductase family 1, member C4 |
| chr16_-_29517141 | 0.22 |
ENST00000550665.1 |
RP11-231C14.4 |
Uncharacterized protein |
| chr2_-_55496174 | 0.22 |
ENST00000417363.1 ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2 |
mitochondrial translational initiation factor 2 |
| chr3_+_52448539 | 0.21 |
ENST00000461861.1 |
PHF7 |
PHD finger protein 7 |
| chr11_-_111794446 | 0.21 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr16_-_15463926 | 0.20 |
ENST00000432570.2 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr1_+_13736907 | 0.20 |
ENST00000316412.5 |
PRAMEF20 |
PRAME family member 20 |
| chr9_-_6645628 | 0.20 |
ENST00000321612.6 |
GLDC |
glycine dehydrogenase (decarboxylating) |
| chr14_+_73563735 | 0.20 |
ENST00000532192.1 |
RBM25 |
RNA binding motif protein 25 |
| chr1_-_109618566 | 0.19 |
ENST00000338366.5 |
TAF13 |
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr1_+_158149737 | 0.19 |
ENST00000368171.3 |
CD1D |
CD1d molecule |
| chr15_-_72523454 | 0.18 |
ENST00000565154.1 ENST00000565184.1 ENST00000389093.3 ENST00000449901.2 ENST00000335181.5 ENST00000319622.6 |
PKM |
pyruvate kinase, muscle |
| chr1_+_13359819 | 0.18 |
ENST00000376168.1 |
PRAMEF5 |
PRAME family member 5 |
| chr21_+_37442239 | 0.18 |
ENST00000530908.1 ENST00000290349.6 ENST00000439427.2 ENST00000399191.3 |
CBR1 |
carbonyl reductase 1 |
| chr4_+_89299994 | 0.18 |
ENST00000264346.7 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr14_-_24911448 | 0.18 |
ENST00000555355.1 ENST00000553343.1 ENST00000556523.1 ENST00000556249.1 ENST00000538105.2 ENST00000555225.1 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
| chr2_-_55496344 | 0.18 |
ENST00000403721.1 ENST00000263629.4 |
MTIF2 |
mitochondrial translational initiation factor 2 |
| chr20_+_34824355 | 0.16 |
ENST00000397286.3 ENST00000320849.4 ENST00000373932.3 |
AAR2 |
AAR2 splicing factor homolog (S. cerevisiae) |
| chr2_+_105953972 | 0.16 |
ENST00000410049.1 |
C2orf49 |
chromosome 2 open reading frame 49 |
| chr3_-_178984759 | 0.16 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr1_-_54665664 | 0.16 |
ENST00000542737.1 ENST00000537208.1 |
CYB5RL |
cytochrome b5 reductase-like |
| chr2_-_219134822 | 0.15 |
ENST00000444053.1 ENST00000248450.4 |
AAMP |
angio-associated, migratory cell protein |
| chr1_+_169337172 | 0.15 |
ENST00000367807.3 ENST00000367808.3 ENST00000329281.2 ENST00000420531.1 |
BLZF1 |
basic leucine zipper nuclear factor 1 |
| chr2_+_61404624 | 0.15 |
ENST00000394457.3 |
AHSA2 |
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr10_+_104613980 | 0.15 |
ENST00000339834.5 |
C10orf32 |
chromosome 10 open reading frame 32 |
| chr11_+_73661364 | 0.15 |
ENST00000339764.1 |
DNAJB13 |
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr16_+_451826 | 0.14 |
ENST00000219481.5 ENST00000397710.1 ENST00000424398.2 |
DECR2 |
2,4-dienoyl CoA reductase 2, peroxisomal |
| chr19_+_36606354 | 0.14 |
ENST00000589996.1 ENST00000591296.1 |
TBCB |
tubulin folding cofactor B |
| chr6_-_35656712 | 0.14 |
ENST00000357266.4 ENST00000542713.1 |
FKBP5 |
FK506 binding protein 5 |
| chr7_+_72349920 | 0.14 |
ENST00000395270.1 ENST00000446813.1 ENST00000257622.4 |
POM121 |
POM121 transmembrane nucleoporin |
| chr11_-_76155618 | 0.14 |
ENST00000530759.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr6_-_31509714 | 0.14 |
ENST00000456662.1 ENST00000431908.1 ENST00000456976.1 ENST00000428450.1 ENST00000453105.2 ENST00000418897.1 ENST00000415382.2 ENST00000449074.2 ENST00000419020.1 ENST00000428098.1 |
DDX39B |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chrX_+_144908928 | 0.14 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr1_+_145293114 | 0.14 |
ENST00000369338.1 |
NBPF10 |
neuroblastoma breakpoint family, member 10 |
| chr16_+_69458537 | 0.14 |
ENST00000515314.1 ENST00000561792.1 ENST00000568237.1 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
| chr13_+_28712614 | 0.13 |
ENST00000380958.3 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr6_-_35656685 | 0.13 |
ENST00000539068.1 ENST00000540787.1 |
FKBP5 |
FK506 binding protein 5 |
| chr14_-_24740709 | 0.13 |
ENST00000399409.3 ENST00000216840.6 |
RABGGTA |
Rab geranylgeranyltransferase, alpha subunit |
| chr14_+_75761099 | 0.13 |
ENST00000561000.1 ENST00000558575.1 |
RP11-293M10.5 |
RP11-293M10.5 |
| chr14_-_21924209 | 0.13 |
ENST00000557364.1 |
CHD8 |
chromodomain helicase DNA binding protein 8 |
| chr16_+_29690358 | 0.13 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr11_-_59633951 | 0.13 |
ENST00000257264.3 |
TCN1 |
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr15_+_62853562 | 0.13 |
ENST00000561311.1 |
TLN2 |
talin 2 |
| chr6_-_167797887 | 0.13 |
ENST00000476779.2 ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10 |
t-complex 10 |
| chr17_-_6915646 | 0.12 |
ENST00000574377.1 ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2 |
Uncharacterized protein |
| chr18_+_616672 | 0.12 |
ENST00000338387.7 |
CLUL1 |
clusterin-like 1 (retinal) |
| chr19_-_11039188 | 0.12 |
ENST00000588347.1 |
YIPF2 |
Yip1 domain family, member 2 |
| chr7_-_73133959 | 0.12 |
ENST00000395155.3 ENST00000395154.3 ENST00000222812.3 ENST00000395156.3 |
STX1A |
syntaxin 1A (brain) |
| chr8_+_119294456 | 0.12 |
ENST00000366457.2 |
AC023590.1 |
Uncharacterized protein |
| chr11_+_117947724 | 0.12 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr2_-_9563319 | 0.12 |
ENST00000497105.1 ENST00000360635.3 ENST00000359712.3 |
ITGB1BP1 |
integrin beta 1 binding protein 1 |
| chr1_+_152975488 | 0.12 |
ENST00000542696.1 |
SPRR3 |
small proline-rich protein 3 |
| chr3_-_9834375 | 0.11 |
ENST00000343450.2 ENST00000301964.2 |
TADA3 |
transcriptional adaptor 3 |
| chr2_-_152589670 | 0.11 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chr20_+_44637526 | 0.11 |
ENST00000372330.3 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr1_+_12916941 | 0.11 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr3_+_70048881 | 0.11 |
ENST00000483525.1 |
RP11-460N16.1 |
RP11-460N16.1 |
| chr10_-_6622258 | 0.11 |
ENST00000263125.5 |
PRKCQ |
protein kinase C, theta |
| chr11_+_117947782 | 0.11 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr3_-_178976996 | 0.11 |
ENST00000485523.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr8_-_134115118 | 0.11 |
ENST00000395352.3 ENST00000338087.5 |
SLA |
Src-like-adaptor |
| chr1_-_53793725 | 0.11 |
ENST00000371454.2 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr1_-_85462623 | 0.10 |
ENST00000370608.3 |
MCOLN2 |
mucolipin 2 |
| chr13_-_41837620 | 0.10 |
ENST00000379477.1 ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1 |
mitochondrial translational release factor 1 |
| chr6_-_11779014 | 0.10 |
ENST00000229583.5 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_+_120049826 | 0.10 |
ENST00000369413.3 ENST00000235547.6 ENST00000528909.1 |
HSD3B1 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 |
| chr6_-_11779174 | 0.10 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr1_-_13007420 | 0.10 |
ENST00000376189.1 |
PRAMEF6 |
PRAME family member 6 |
| chr19_-_15344243 | 0.10 |
ENST00000602233.1 |
EPHX3 |
epoxide hydrolase 3 |
| chr11_-_76155700 | 0.10 |
ENST00000572035.1 |
RP11-111M22.3 |
RP11-111M22.3 |
| chr10_-_74856608 | 0.10 |
ENST00000307116.2 ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1 |
prolyl 4-hydroxylase, alpha polypeptide I |
| chr12_-_118796910 | 0.10 |
ENST00000541186.1 ENST00000539872.1 |
TAOK3 |
TAO kinase 3 |
| chr9_+_6716478 | 0.09 |
ENST00000452643.1 |
RP11-390F4.3 |
RP11-390F4.3 |
| chr14_-_95942173 | 0.09 |
ENST00000334258.5 ENST00000557275.1 ENST00000553340.1 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
| chr6_-_52710893 | 0.09 |
ENST00000284562.2 |
GSTA5 |
glutathione S-transferase alpha 5 |
| chr22_+_32455111 | 0.09 |
ENST00000543737.1 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr12_-_96429423 | 0.09 |
ENST00000228740.2 |
LTA4H |
leukotriene A4 hydrolase |
| chr4_-_153332886 | 0.08 |
ENST00000603841.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_160709076 | 0.08 |
ENST00000359331.4 ENST00000495334.1 |
SLAMF7 |
SLAM family member 7 |
| chr8_-_134072593 | 0.08 |
ENST00000427060.2 |
SLA |
Src-like-adaptor |
| chr1_+_215740709 | 0.08 |
ENST00000259154.4 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
| chr1_-_13115578 | 0.08 |
ENST00000414205.2 |
PRAMEF6 |
PRAME family member 6 |
| chr15_+_40331456 | 0.08 |
ENST00000504245.1 ENST00000560341.1 |
SRP14-AS1 |
SRP14 antisense RNA1 (head to head) |
| chr21_-_34185944 | 0.08 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr1_-_13117736 | 0.08 |
ENST00000376192.5 ENST00000376182.1 |
PRAMEF6 |
PRAME family member 6 |
| chr3_+_89156674 | 0.08 |
ENST00000336596.2 |
EPHA3 |
EPH receptor A3 |
| chr13_+_53226963 | 0.08 |
ENST00000343788.6 ENST00000535397.1 ENST00000310528.8 |
SUGT1 |
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr1_-_150979333 | 0.08 |
ENST00000312210.5 |
FAM63A |
family with sequence similarity 63, member A |
| chr7_-_71912046 | 0.08 |
ENST00000395276.2 ENST00000431984.1 |
CALN1 |
calneuron 1 |
| chr7_+_12727250 | 0.08 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr11_+_27062502 | 0.08 |
ENST00000263182.3 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr1_-_13390765 | 0.08 |
ENST00000357367.2 |
PRAMEF8 |
PRAME family member 8 |
| chr19_+_57999101 | 0.07 |
ENST00000347466.6 ENST00000523138.1 ENST00000415379.2 ENST00000521754.1 ENST00000221735.7 ENST00000518999.1 ENST00000521137.1 |
ZNF419 |
zinc finger protein 419 |
| chr4_-_84255935 | 0.07 |
ENST00000513463.1 |
HPSE |
heparanase |
| chr1_+_244816237 | 0.07 |
ENST00000302550.11 |
DESI2 |
desumoylating isopeptidase 2 |
| chr7_-_14880892 | 0.07 |
ENST00000406247.3 ENST00000399322.3 ENST00000258767.5 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr5_-_59481406 | 0.07 |
ENST00000546160.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr3_+_149191723 | 0.07 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr19_+_1026298 | 0.07 |
ENST00000263097.4 |
CNN2 |
calponin 2 |
| chr18_-_51750948 | 0.07 |
ENST00000583046.1 ENST00000398398.2 |
MBD2 |
methyl-CpG binding domain protein 2 |
| chr20_+_18488137 | 0.07 |
ENST00000450074.1 ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
| chr5_+_70220768 | 0.07 |
ENST00000351205.4 ENST00000503079.2 ENST00000380707.4 ENST00000514951.1 ENST00000506163.1 |
SMN1 |
survival of motor neuron 1, telomeric |
| chr2_+_161993465 | 0.07 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr6_+_31802364 | 0.07 |
ENST00000375640.3 ENST00000375641.2 |
C6orf48 |
chromosome 6 open reading frame 48 |
| chr8_+_18248755 | 0.07 |
ENST00000286479.3 |
NAT2 |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr2_-_127963343 | 0.07 |
ENST00000335247.7 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr16_+_16481306 | 0.06 |
ENST00000422673.2 |
NPIPA7 |
nuclear pore complex interacting protein family, member A7 |
| chr2_+_228189867 | 0.06 |
ENST00000423098.1 ENST00000304593.9 |
MFF |
mitochondrial fission factor |
| chr19_+_57999079 | 0.06 |
ENST00000426954.2 ENST00000354197.4 ENST00000523882.1 ENST00000520540.1 ENST00000519310.1 ENST00000442920.2 ENST00000523312.1 ENST00000424930.2 |
ZNF419 |
zinc finger protein 419 |
| chr1_+_186265399 | 0.06 |
ENST00000367486.3 ENST00000367484.3 ENST00000533951.1 ENST00000367482.4 ENST00000367483.4 ENST00000367485.4 ENST00000445192.2 |
PRG4 |
proteoglycan 4 |
| chr1_-_43637915 | 0.06 |
ENST00000236051.2 |
EBNA1BP2 |
EBNA1 binding protein 2 |
| chr4_+_70894130 | 0.06 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr8_+_18248786 | 0.06 |
ENST00000520116.1 |
NAT2 |
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr7_+_16685756 | 0.06 |
ENST00000415365.1 ENST00000258761.3 ENST00000433922.2 ENST00000452975.2 ENST00000405202.1 |
BZW2 |
basic leucine zipper and W2 domains 2 |
| chr19_+_37464063 | 0.06 |
ENST00000586353.1 ENST00000433993.1 ENST00000592567.1 |
ZNF568 |
zinc finger protein 568 |
| chr4_-_83719884 | 0.06 |
ENST00000282709.4 ENST00000273908.4 |
SCD5 |
stearoyl-CoA desaturase 5 |
| chr2_-_158732340 | 0.06 |
ENST00000539637.1 ENST00000413751.1 ENST00000434821.1 ENST00000424669.1 |
ACVR1 |
activin A receptor, type I |
| chr5_-_81574160 | 0.06 |
ENST00000510210.1 ENST00000512493.1 ENST00000507980.1 ENST00000511844.1 ENST00000510019.1 |
RPS23 |
ribosomal protein S23 |
| chr1_-_12958101 | 0.05 |
ENST00000235347.4 |
PRAMEF10 |
PRAME family member 10 |
| chr12_-_10962767 | 0.05 |
ENST00000240691.2 |
TAS2R9 |
taste receptor, type 2, member 9 |
| chr1_+_12976450 | 0.05 |
ENST00000361079.2 |
PRAMEF7 |
PRAME family member 7 |
| chr20_-_44600810 | 0.05 |
ENST00000322927.2 ENST00000426788.1 |
ZNF335 |
zinc finger protein 335 |
| chr22_+_41074754 | 0.05 |
ENST00000249016.4 |
MCHR1 |
melanin-concentrating hormone receptor 1 |
| chr17_+_37356586 | 0.05 |
ENST00000579260.1 ENST00000582193.1 |
RPL19 |
ribosomal protein L19 |
| chr19_-_48547294 | 0.05 |
ENST00000293255.2 |
CABP5 |
calcium binding protein 5 |
| chr3_+_35722487 | 0.05 |
ENST00000441454.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chrX_-_65259900 | 0.05 |
ENST00000412866.2 |
VSIG4 |
V-set and immunoglobulin domain containing 4 |
| chr17_-_56769382 | 0.05 |
ENST00000240361.8 ENST00000349033.5 ENST00000389934.3 |
TEX14 |
testis expressed 14 |
| chrX_+_1733876 | 0.05 |
ENST00000381241.3 |
ASMT |
acetylserotonin O-methyltransferase |
| chr15_-_90892669 | 0.05 |
ENST00000412799.2 |
GABARAPL3 |
GABA(A) receptors associated protein like 3, pseudogene |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.6 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.4 | 1.2 | GO:1900138 | negative regulation of phospholipase A2 activity(GO:1900138) |
| 0.2 | 0.7 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.2 | 1.7 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.2 | 1.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 1.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.2 | 0.5 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.2 | 2.7 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.6 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 0.5 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.6 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 1.1 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.1 | 0.8 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.3 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.3 | GO:1902023 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.1 | 0.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.1 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.1 | 0.6 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.4 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.2 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:2000570 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.9 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.0 | 0.2 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.0 | 0.2 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 1.3 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.0 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0052422 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) negative regulation by host of symbiont molecular function(GO:0052405) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0097180 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.8 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.1 | 0.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.2 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 2.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.3 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.0 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.4 | 1.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 4.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.8 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.5 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.4 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.6 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 2.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.1 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |