Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for SIX6
Z-value: 1.07
Transcription factors associated with SIX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX6
|
ENSG00000184302.6 | SIX6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SIX6 | hg19_v2_chr14_+_60975644_60975673 | 0.12 | 6.7e-01 | Click! |
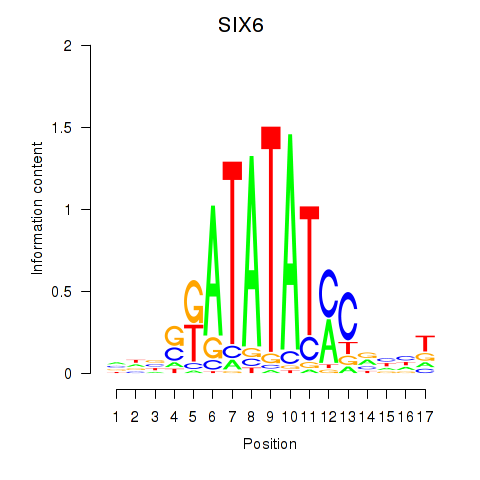
Activity profile of SIX6 motif
Sorted Z-values of SIX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SIX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_88427568 | 4.97 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chr1_-_161193349 | 4.29 |
ENST00000469730.2 ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2 |
apolipoprotein A-II |
| chr4_-_155511887 | 2.85 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chr4_+_74301880 | 2.44 |
ENST00000395792.2 ENST00000226359.2 |
AFP |
alpha-fetoprotein |
| chr13_-_46679185 | 2.35 |
ENST00000439329.3 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr13_-_46679144 | 2.01 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr20_+_34802295 | 1.82 |
ENST00000432603.1 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
| chr16_+_72090053 | 1.59 |
ENST00000576168.2 ENST00000567185.3 ENST00000567612.2 |
HP |
haptoglobin |
| chr4_-_71532339 | 1.23 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr1_+_160709055 | 1.16 |
ENST00000368043.3 ENST00000368042.3 ENST00000458602.2 ENST00000458104.2 |
SLAMF7 |
SLAM family member 7 |
| chr14_+_21152259 | 1.10 |
ENST00000555835.1 ENST00000336811.6 |
RNASE4 ANG |
ribonuclease, RNase A family, 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr12_-_122018859 | 1.07 |
ENST00000536437.1 ENST00000377071.4 ENST00000538046.2 |
KDM2B |
lysine (K)-specific demethylase 2B |
| chr15_-_80263506 | 1.07 |
ENST00000335661.6 |
BCL2A1 |
BCL2-related protein A1 |
| chr12_-_28123206 | 1.01 |
ENST00000542963.1 ENST00000535992.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr1_+_160709029 | 0.98 |
ENST00000444090.2 ENST00000441662.2 |
SLAMF7 |
SLAM family member 7 |
| chrX_-_20236970 | 0.90 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr12_-_28122980 | 0.89 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr8_+_101170257 | 0.77 |
ENST00000251809.3 |
SPAG1 |
sperm associated antigen 1 |
| chr16_+_22524844 | 0.76 |
ENST00000538606.1 ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr6_+_26240561 | 0.72 |
ENST00000377745.2 |
HIST1H4F |
histone cluster 1, H4f |
| chr3_-_9994021 | 0.66 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr1_+_156119798 | 0.64 |
ENST00000355014.2 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr19_+_3136115 | 0.64 |
ENST00000262958.3 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
| chr16_-_21868978 | 0.61 |
ENST00000357370.5 ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr16_-_21436459 | 0.58 |
ENST00000448012.2 ENST00000504841.2 ENST00000419180.2 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr19_-_14628645 | 0.56 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr2_-_58468437 | 0.56 |
ENST00000403676.1 ENST00000427708.2 ENST00000403295.3 ENST00000446381.1 ENST00000417361.1 ENST00000233741.4 ENST00000402135.3 ENST00000540646.1 ENST00000449070.1 |
FANCL |
Fanconi anemia, complementation group L |
| chr1_-_67142710 | 0.55 |
ENST00000502413.2 |
AL139147.1 |
Uncharacterized protein |
| chr21_-_43735446 | 0.51 |
ENST00000398431.2 |
TFF3 |
trefoil factor 3 (intestinal) |
| chr12_-_498620 | 0.49 |
ENST00000399788.2 ENST00000382815.4 |
KDM5A |
lysine (K)-specific demethylase 5A |
| chr16_-_21868739 | 0.42 |
ENST00000415645.2 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chrX_+_12809463 | 0.41 |
ENST00000380663.3 ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2 |
phosphoribosyl pyrophosphate synthetase 2 |
| chr20_-_4990931 | 0.41 |
ENST00000379333.1 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr3_-_111314230 | 0.40 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr19_-_16008880 | 0.39 |
ENST00000011989.7 ENST00000221700.6 |
CYP4F2 |
cytochrome P450, family 4, subfamily F, polypeptide 2 |
| chr9_-_99382065 | 0.37 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr12_-_46385811 | 0.36 |
ENST00000419565.2 |
SCAF11 |
SR-related CTD-associated factor 11 |
| chr10_-_61720640 | 0.36 |
ENST00000521074.1 ENST00000444900.1 |
C10orf40 |
chromosome 10 open reading frame 40 |
| chr1_-_150669604 | 0.35 |
ENST00000427665.1 ENST00000540514.1 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr2_-_208989225 | 0.35 |
ENST00000264376.4 |
CRYGD |
crystallin, gamma D |
| chr1_+_53308398 | 0.34 |
ENST00000371528.1 |
ZYG11A |
zyg-11 family member A, cell cycle regulator |
| chr16_-_29517141 | 0.33 |
ENST00000550665.1 |
RP11-231C14.4 |
Uncharacterized protein |
| chr3_+_151531810 | 0.32 |
ENST00000232892.7 |
AADAC |
arylacetamide deacetylase |
| chr5_+_140201183 | 0.32 |
ENST00000529619.1 ENST00000529859.1 ENST00000378126.3 |
PCDHA5 |
protocadherin alpha 5 |
| chr17_-_5342380 | 0.31 |
ENST00000225698.4 |
C1QBP |
complement component 1, q subcomponent binding protein |
| chr21_-_43735628 | 0.28 |
ENST00000291525.10 ENST00000518498.1 |
TFF3 |
trefoil factor 3 (intestinal) |
| chrX_-_18238999 | 0.27 |
ENST00000380033.4 ENST00000380030.3 |
BEND2 |
BEN domain containing 2 |
| chr2_+_89184868 | 0.27 |
ENST00000390243.2 |
IGKV4-1 |
immunoglobulin kappa variable 4-1 |
| chr2_-_77749474 | 0.27 |
ENST00000409093.1 ENST00000409088.3 |
LRRTM4 |
leucine rich repeat transmembrane neuronal 4 |
| chr2_-_89266286 | 0.26 |
ENST00000464162.1 |
IGKV1-6 |
immunoglobulin kappa variable 1-6 |
| chr11_+_74699942 | 0.26 |
ENST00000526068.1 ENST00000532963.1 ENST00000531619.1 ENST00000534628.1 ENST00000545272.1 |
NEU3 |
sialidase 3 (membrane sialidase) |
| chr6_+_26204825 | 0.25 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chr1_+_212738676 | 0.24 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr16_+_28648975 | 0.23 |
ENST00000529716.1 |
NPIPB8 |
nuclear pore complex interacting protein family, member B8 |
| chr17_-_42327236 | 0.23 |
ENST00000399246.2 |
AC003102.1 |
AC003102.1 |
| chr5_+_40909354 | 0.23 |
ENST00000313164.9 |
C7 |
complement component 7 |
| chr2_+_90192768 | 0.22 |
ENST00000390275.2 |
IGKV1D-13 |
immunoglobulin kappa variable 1D-13 |
| chr13_+_73629107 | 0.21 |
ENST00000539231.1 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr17_+_60501228 | 0.20 |
ENST00000311506.5 |
METTL2A |
methyltransferase like 2A |
| chr6_-_26189304 | 0.20 |
ENST00000340756.2 |
HIST1H4D |
histone cluster 1, H4d |
| chr1_-_157014865 | 0.20 |
ENST00000361409.2 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr13_+_34392185 | 0.20 |
ENST00000380071.3 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
| chr9_-_21351377 | 0.19 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chr22_-_19137796 | 0.19 |
ENST00000086933.2 |
GSC2 |
goosecoid homeobox 2 |
| chr2_-_62081148 | 0.18 |
ENST00000404929.1 |
FAM161A |
family with sequence similarity 161, member A |
| chr12_-_87232644 | 0.18 |
ENST00000549405.2 |
MGAT4C |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr15_-_51397473 | 0.17 |
ENST00000327536.5 |
TNFAIP8L3 |
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chrX_+_38660704 | 0.16 |
ENST00000378474.3 |
MID1IP1 |
MID1 interacting protein 1 |
| chrX_+_64708615 | 0.16 |
ENST00000338957.4 ENST00000423889.3 |
ZC3H12B |
zinc finger CCCH-type containing 12B |
| chr11_+_33902189 | 0.15 |
ENST00000330381.2 |
AC132216.1 |
HCG1785179; PRO1787; Uncharacterized protein |
| chr5_-_66492562 | 0.15 |
ENST00000256447.4 |
CD180 |
CD180 molecule |
| chr19_+_55235969 | 0.15 |
ENST00000402254.2 ENST00000538269.1 ENST00000541392.1 ENST00000291860.1 ENST00000396284.2 |
KIR3DL1 KIR3DL3 KIR2DL4 |
killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 1 killer cell immunoglobulin-like receptor, three domains, long cytoplasmic tail, 3 killer cell immunoglobulin-like receptor, two domains, long cytoplasmic tail, 4 |
| chr11_-_102496063 | 0.12 |
ENST00000260228.2 |
MMP20 |
matrix metallopeptidase 20 |
| chrY_-_24038660 | 0.12 |
ENST00000382677.3 |
RBMY1D |
RNA binding motif protein, Y-linked, family 1, member D |
| chr11_-_45307817 | 0.12 |
ENST00000020926.3 |
SYT13 |
synaptotagmin XIII |
| chr11_+_122709200 | 0.12 |
ENST00000227348.4 |
CRTAM |
cytotoxic and regulatory T cell molecule |
| chr8_-_141774467 | 0.12 |
ENST00000520151.1 ENST00000519024.1 ENST00000519465.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr6_-_26285737 | 0.11 |
ENST00000377727.1 ENST00000289352.1 |
HIST1H4H |
histone cluster 1, H4h |
| chrX_+_8433376 | 0.11 |
ENST00000440654.2 ENST00000381029.4 |
VCX3B |
variable charge, X-linked 3B |
| chr10_+_52094298 | 0.10 |
ENST00000595931.1 |
AC069547.1 |
HCG1745369; PRO3073; Uncharacterized protein |
| chr6_+_52051171 | 0.10 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chrY_+_23698778 | 0.08 |
ENST00000303902.5 |
RBMY1A1 |
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr6_+_146348810 | 0.08 |
ENST00000492807.2 |
GRM1 |
glutamate receptor, metabotropic 1 |
| chr12_-_121477039 | 0.07 |
ENST00000257570.5 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr4_-_123377880 | 0.07 |
ENST00000226730.4 |
IL2 |
interleukin 2 |
| chr16_-_20364030 | 0.07 |
ENST00000396134.2 ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD |
uromodulin |
| chr16_+_19467772 | 0.07 |
ENST00000219821.5 ENST00000561503.1 ENST00000564959.1 |
TMC5 |
transmembrane channel-like 5 |
| chr8_+_1993173 | 0.06 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr6_+_146348782 | 0.06 |
ENST00000361719.2 ENST00000392299.2 |
GRM1 |
glutamate receptor, metabotropic 1 |
| chr16_-_20364122 | 0.06 |
ENST00000396138.4 ENST00000577168.1 |
UMOD |
uromodulin |
| chr2_+_177134134 | 0.05 |
ENST00000249442.6 ENST00000392529.2 ENST00000443241.1 |
MTX2 |
metaxin 2 |
| chr2_+_177134201 | 0.05 |
ENST00000452865.1 |
MTX2 |
metaxin 2 |
| chr12_-_10978957 | 0.04 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chrX_-_55020511 | 0.04 |
ENST00000375006.3 ENST00000374992.2 |
PFKFB1 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr1_+_159557607 | 0.04 |
ENST00000255040.2 |
APCS |
amyloid P component, serum |
| chr12_-_121476750 | 0.04 |
ENST00000543677.1 |
OASL |
2'-5'-oligoadenylate synthetase-like |
| chr6_-_26018007 | 0.04 |
ENST00000244573.3 |
HIST1H1A |
histone cluster 1, H1a |
| chr4_+_88532028 | 0.03 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chrX_+_128674213 | 0.02 |
ENST00000371113.4 ENST00000357121.5 |
OCRL |
oculocerebrorenal syndrome of Lowe |
| chrX_-_118699325 | 0.02 |
ENST00000320339.4 ENST00000371594.4 ENST00000536133.1 |
CXorf56 |
chromosome X open reading frame 56 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.3 | GO:0060621 | regulation of cholesterol import(GO:0060620) negative regulation of cholesterol import(GO:0060621) regulation of sterol import(GO:2000909) negative regulation of sterol import(GO:2000910) |
| 1.1 | 4.4 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.5 | 1.6 | GO:2000296 | negative regulation of hydrogen peroxide catabolic process(GO:2000296) |
| 0.5 | 5.0 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.4 | 1.1 | GO:0021678 | midbrain-hindbrain boundary morphogenesis(GO:0021555) fourth ventricle development(GO:0021592) third ventricle development(GO:0021678) |
| 0.3 | 2.9 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.2 | 1.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 2.4 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.1 | 0.4 | GO:0003095 | pressure natriuresis(GO:0003095) negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.5 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.6 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.4 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.2 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.1 | 0.3 | GO:1901165 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.8 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.4 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.6 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 1.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.3 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.0 | 0.4 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.9 | GO:0043555 | regulation of translation in response to stress(GO:0043555) |
| 0.0 | 0.2 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 2.3 | GO:0042267 | natural killer cell mediated cytotoxicity(GO:0042267) |
| 0.0 | 0.4 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.1 | GO:2000320 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.0 | 0.2 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.6 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 2.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.8 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 1.1 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.8 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.7 | 5.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 1.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 4.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.4 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.4 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.5 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.3 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 1.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 1.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.0 | GO:0045179 | apical cortex(GO:0045179) |
| 0.4 | 1.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.3 | 4.3 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.2 | 2.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.2 | 1.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 0.2 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.2 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.9 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 5.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.8 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.2 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |