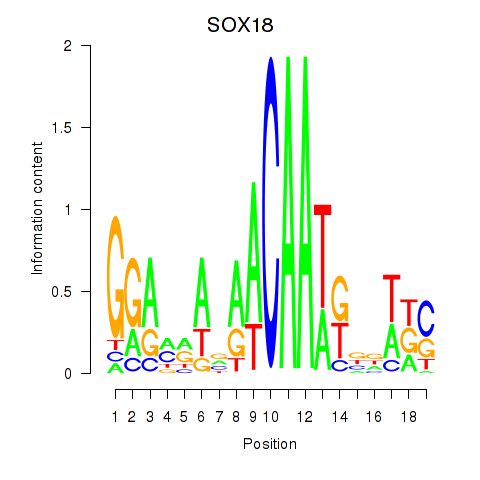
Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for SOX18
Z-value: 0.80
Transcription factors associated with SOX18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX18
|
ENSG00000203883.5 | SOX18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX18 | hg19_v2_chr20_-_62680984_62680999 | 0.47 | 6.4e-02 | Click! |
Activity profile of SOX18 motif
Sorted Z-values of SOX18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_15918618 | 2.11 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr1_-_32801825 | 1.85 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chrX_+_30261847 | 1.41 |
ENST00000378981.3 ENST00000397550.1 |
MAGEB1 |
melanoma antigen family B, 1 |
| chr13_-_46756351 | 1.27 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr13_-_46716969 | 1.25 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr3_-_121379739 | 1.23 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr7_-_150329421 | 1.23 |
ENST00000493969.1 ENST00000328902.5 |
GIMAP6 |
GTPase, IMAP family member 6 |
| chrX_-_154033661 | 0.99 |
ENST00000393531.1 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
| chr11_-_89224488 | 0.93 |
ENST00000534731.1 ENST00000527626.1 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89224508 | 0.88 |
ENST00000525196.1 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89224299 | 0.86 |
ENST00000343727.5 ENST00000531342.1 ENST00000375979.3 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89223883 | 0.80 |
ENST00000528341.1 |
NOX4 |
NADPH oxidase 4 |
| chr11_-_89224139 | 0.79 |
ENST00000413594.2 |
NOX4 |
NADPH oxidase 4 |
| chr2_+_170590321 | 0.78 |
ENST00000392647.2 |
KLHL23 |
kelch-like family member 23 |
| chr4_-_144940477 | 0.78 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr2_-_158345462 | 0.76 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr6_-_26235206 | 0.74 |
ENST00000244534.5 |
HIST1H1D |
histone cluster 1, H1d |
| chr15_-_55541227 | 0.73 |
ENST00000566877.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr4_+_154387480 | 0.68 |
ENST00000409663.3 ENST00000440693.1 ENST00000409959.3 |
KIAA0922 |
KIAA0922 |
| chrX_-_70329118 | 0.65 |
ENST00000374188.3 |
IL2RG |
interleukin 2 receptor, gamma |
| chr6_-_26216872 | 0.64 |
ENST00000244601.3 |
HIST1H2BG |
histone cluster 1, H2bg |
| chr11_-_5248294 | 0.64 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr8_-_86253888 | 0.57 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr19_-_46105411 | 0.56 |
ENST00000323040.4 ENST00000544371.1 |
GPR4 OPA3 |
G protein-coupled receptor 4 optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
| chr1_+_100316041 | 0.55 |
ENST00000370165.3 ENST00000370163.3 ENST00000294724.4 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| chr12_-_772901 | 0.55 |
ENST00000305108.4 |
NINJ2 |
ninjurin 2 |
| chr1_-_155880672 | 0.54 |
ENST00000609492.1 ENST00000368322.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr3_+_133465228 | 0.51 |
ENST00000482271.1 ENST00000264998.3 |
TF |
transferrin |
| chr8_+_40010989 | 0.51 |
ENST00000315792.3 |
C8orf4 |
chromosome 8 open reading frame 4 |
| chr1_+_66258846 | 0.50 |
ENST00000341517.4 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr18_+_32621324 | 0.47 |
ENST00000300249.5 ENST00000538170.2 ENST00000588910.1 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr15_-_68497657 | 0.45 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr1_+_43124087 | 0.43 |
ENST00000304979.3 ENST00000372550.1 ENST00000440068.1 |
PPIH |
peptidylprolyl isomerase H (cyclophilin H) |
| chr6_+_26217159 | 0.43 |
ENST00000303910.2 |
HIST1H2AE |
histone cluster 1, H2ae |
| chr11_-_6677018 | 0.43 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr14_-_71107921 | 0.40 |
ENST00000553982.1 ENST00000500016.1 |
CTD-2540L5.5 CTD-2540L5.6 |
CTD-2540L5.5 CTD-2540L5.6 |
| chrX_-_101726732 | 0.40 |
ENST00000457521.2 ENST00000412230.2 ENST00000453326.2 |
NXF2B TCP11X2 |
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
| chr8_-_37707356 | 0.39 |
ENST00000520601.1 ENST00000521170.1 ENST00000220659.6 |
BRF2 |
BRF2, RNA polymerase III transcription initiation factor 50 kDa subunit |
| chrX_+_101470280 | 0.38 |
ENST00000395088.2 ENST00000330252.5 ENST00000333110.5 |
NXF2 TCP11X1 |
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr4_+_109541772 | 0.38 |
ENST00000506397.1 ENST00000394668.2 |
RPL34 |
ribosomal protein L34 |
| chr22_-_22292934 | 0.37 |
ENST00000538191.1 ENST00000424647.1 ENST00000407142.1 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr14_-_107219365 | 0.36 |
ENST00000424969.2 |
IGHV3-74 |
immunoglobulin heavy variable 3-74 |
| chr19_+_33571786 | 0.35 |
ENST00000170564.2 |
GPATCH1 |
G patch domain containing 1 |
| chr1_-_182361327 | 0.35 |
ENST00000331872.6 ENST00000311223.5 |
GLUL |
glutamate-ammonia ligase |
| chr8_+_38644778 | 0.34 |
ENST00000276520.8 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr13_+_111806083 | 0.34 |
ENST00000375736.4 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr12_+_96252706 | 0.34 |
ENST00000266735.5 ENST00000553192.1 ENST00000552085.1 |
SNRPF |
small nuclear ribonucleoprotein polypeptide F |
| chr5_+_31532373 | 0.33 |
ENST00000325366.9 ENST00000355907.3 ENST00000507818.2 |
C5orf22 |
chromosome 5 open reading frame 22 |
| chr1_-_11918988 | 0.32 |
ENST00000376468.3 |
NPPB |
natriuretic peptide B |
| chr1_-_182360918 | 0.32 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chr6_-_13487784 | 0.31 |
ENST00000379287.3 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
| chr8_-_27469196 | 0.31 |
ENST00000546343.1 ENST00000560566.1 |
CLU |
clusterin |
| chr5_+_32585605 | 0.30 |
ENST00000265073.4 ENST00000515355.1 ENST00000502897.1 ENST00000510442.1 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
| chr1_+_36348790 | 0.30 |
ENST00000373204.4 |
AGO1 |
argonaute RISC catalytic component 1 |
| chr1_-_241799232 | 0.29 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr3_+_186330712 | 0.28 |
ENST00000411641.2 ENST00000273784.5 |
AHSG |
alpha-2-HS-glycoprotein |
| chr8_-_27468842 | 0.27 |
ENST00000523500.1 |
CLU |
clusterin |
| chr5_+_44809027 | 0.27 |
ENST00000507110.1 |
MRPS30 |
mitochondrial ribosomal protein S30 |
| chr5_-_55412774 | 0.26 |
ENST00000434982.2 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr4_+_109541740 | 0.26 |
ENST00000394665.1 |
RPL34 |
ribosomal protein L34 |
| chr6_-_32908792 | 0.25 |
ENST00000418107.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr4_+_109541722 | 0.25 |
ENST00000394667.3 ENST00000502534.1 |
RPL34 |
ribosomal protein L34 |
| chr1_+_209929377 | 0.25 |
ENST00000400959.3 ENST00000367025.3 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr1_-_206785789 | 0.25 |
ENST00000437518.1 ENST00000367114.3 |
EIF2D |
eukaryotic translation initiation factor 2D |
| chr1_-_206785898 | 0.24 |
ENST00000271764.2 |
EIF2D |
eukaryotic translation initiation factor 2D |
| chr18_-_5396271 | 0.24 |
ENST00000579951.1 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
| chr10_+_70480963 | 0.23 |
ENST00000265872.6 ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
| chr12_+_15125954 | 0.23 |
ENST00000266395.2 |
PDE6H |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr9_-_74525658 | 0.22 |
ENST00000333421.6 |
ABHD17B |
abhydrolase domain containing 17B |
| chr18_+_74240610 | 0.22 |
ENST00000578092.1 ENST00000578613.1 ENST00000583578.1 |
LINC00908 |
long intergenic non-protein coding RNA 908 |
| chr5_-_31532160 | 0.22 |
ENST00000511367.2 ENST00000513349.1 |
DROSHA |
drosha, ribonuclease type III |
| chr11_+_20620946 | 0.22 |
ENST00000525748.1 |
SLC6A5 |
solute carrier family 6 (neurotransmitter transporter), member 5 |
| chr4_+_69962185 | 0.22 |
ENST00000305231.7 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr19_+_8117636 | 0.21 |
ENST00000253451.4 ENST00000315626.4 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr4_+_69962212 | 0.21 |
ENST00000508661.1 |
UGT2B7 |
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr2_+_105953972 | 0.21 |
ENST00000410049.1 |
C2orf49 |
chromosome 2 open reading frame 49 |
| chr2_-_153573965 | 0.20 |
ENST00000448428.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr22_-_31324215 | 0.20 |
ENST00000429468.1 |
MORC2 |
MORC family CW-type zinc finger 2 |
| chr11_-_75236867 | 0.20 |
ENST00000376282.3 ENST00000336898.3 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr1_-_156828810 | 0.19 |
ENST00000368195.3 |
INSRR |
insulin receptor-related receptor |
| chr2_+_241544834 | 0.19 |
ENST00000319838.5 ENST00000403859.1 ENST00000438013.2 |
GPR35 |
G protein-coupled receptor 35 |
| chr12_-_8088871 | 0.19 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr21_+_30671690 | 0.19 |
ENST00000399921.1 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr12_+_25205446 | 0.19 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr9_-_123812542 | 0.18 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr4_+_54243798 | 0.18 |
ENST00000337488.6 ENST00000358575.5 ENST00000507922.1 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
| chr2_+_87135076 | 0.18 |
ENST00000409776.2 |
RGPD1 |
RANBP2-like and GRIP domain containing 1 |
| chr2_+_217277137 | 0.18 |
ENST00000430374.1 ENST00000357276.4 ENST00000444508.1 |
SMARCAL1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr12_-_71148357 | 0.18 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr3_+_20081515 | 0.18 |
ENST00000263754.4 |
KAT2B |
K(lysine) acetyltransferase 2B |
| chr12_-_71148413 | 0.17 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chrX_+_100645812 | 0.17 |
ENST00000427805.2 ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A RPL36A-HNRNPH2 |
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr19_+_8117881 | 0.17 |
ENST00000390669.3 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr11_+_18230685 | 0.15 |
ENST00000340135.3 ENST00000534640.1 |
RP11-113D6.10 |
Putative mitochondrial carrier protein LOC494141 |
| chr16_-_787771 | 0.15 |
ENST00000568545.1 |
NARFL |
nuclear prelamin A recognition factor-like |
| chr16_+_33629600 | 0.15 |
ENST00000562905.2 |
IGHV3OR16-13 |
immunoglobulin heavy variable 3/OR16-13 (non-functional) |
| chr1_+_209929494 | 0.14 |
ENST00000367026.3 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
| chr10_-_13390270 | 0.14 |
ENST00000378614.4 ENST00000545675.1 ENST00000327347.5 |
SEPHS1 |
selenophosphate synthetase 1 |
| chr6_-_75953484 | 0.14 |
ENST00000472311.2 ENST00000460985.1 ENST00000377978.3 ENST00000509698.1 ENST00000230459.4 ENST00000370089.2 |
COX7A2 |
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr16_-_4896205 | 0.14 |
ENST00000589389.1 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chr19_+_57742431 | 0.14 |
ENST00000302804.7 |
AURKC |
aurora kinase C |
| chr17_+_25799008 | 0.14 |
ENST00000583370.1 ENST00000398988.3 ENST00000268763.6 |
KSR1 |
kinase suppressor of ras 1 |
| chr1_-_153513170 | 0.13 |
ENST00000368717.2 |
S100A5 |
S100 calcium binding protein A5 |
| chr11_+_134201911 | 0.13 |
ENST00000389881.3 |
GLB1L2 |
galactosidase, beta 1-like 2 |
| chr7_-_142176790 | 0.13 |
ENST00000390369.2 |
TRBV7-4 |
T cell receptor beta variable 7-4 (gene/pseudogene) |
| chr7_-_6048650 | 0.12 |
ENST00000382321.4 ENST00000406569.3 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr4_+_110736659 | 0.12 |
ENST00000394631.3 ENST00000226796.6 |
GAR1 |
GAR1 ribonucleoprotein |
| chr17_+_41150479 | 0.12 |
ENST00000589913.1 |
RPL27 |
ribosomal protein L27 |
| chr8_-_102218292 | 0.12 |
ENST00000518336.1 ENST00000520454.1 |
ZNF706 |
zinc finger protein 706 |
| chr11_+_60970852 | 0.12 |
ENST00000325558.6 |
PGA3 |
pepsinogen 3, group I (pepsinogen A) |
| chr12_+_113344582 | 0.11 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr4_-_76944621 | 0.11 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr5_-_75919217 | 0.11 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr1_-_155881156 | 0.11 |
ENST00000539040.1 ENST00000368323.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr15_+_91473403 | 0.10 |
ENST00000394275.2 |
UNC45A |
unc-45 homolog A (C. elegans) |
| chr3_+_108541545 | 0.10 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr9_-_74525847 | 0.10 |
ENST00000377041.2 |
ABHD17B |
abhydrolase domain containing 17B |
| chr2_+_217277466 | 0.10 |
ENST00000358207.5 ENST00000434435.1 |
SMARCAL1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr12_-_371994 | 0.09 |
ENST00000343164.4 ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13 |
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr7_+_12727250 | 0.09 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr2_-_153573887 | 0.09 |
ENST00000493468.2 ENST00000545856.1 |
PRPF40A |
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr6_-_160679905 | 0.09 |
ENST00000366953.3 |
SLC22A2 |
solute carrier family 22 (organic cation transporter), member 2 |
| chr17_+_41158742 | 0.09 |
ENST00000415816.2 ENST00000438323.2 |
IFI35 |
interferon-induced protein 35 |
| chr19_-_7936344 | 0.09 |
ENST00000599142.1 |
CTD-3193O13.9 |
Protein FLJ22184 |
| chr15_+_75628419 | 0.08 |
ENST00000567377.1 ENST00000562789.1 ENST00000568301.1 |
COMMD4 |
COMM domain containing 4 |
| chrX_+_119737806 | 0.08 |
ENST00000371317.5 |
MCTS1 |
malignant T cell amplified sequence 1 |
| chr16_+_24549014 | 0.08 |
ENST00000564314.1 ENST00000567686.1 |
RBBP6 |
retinoblastoma binding protein 6 |
| chr12_+_69633317 | 0.08 |
ENST00000435070.2 |
CPSF6 |
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_+_69633407 | 0.08 |
ENST00000551516.1 |
CPSF6 |
cleavage and polyadenylation specific factor 6, 68kDa |
| chr5_-_114598548 | 0.08 |
ENST00000379615.3 ENST00000419445.1 |
PGGT1B |
protein geranylgeranyltransferase type I, beta subunit |
| chr5_-_78808617 | 0.08 |
ENST00000282260.6 ENST00000508576.1 ENST00000535690.1 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr7_-_93204033 | 0.08 |
ENST00000359558.2 ENST00000360249.4 ENST00000426151.1 |
CALCR |
calcitonin receptor |
| chrX_+_119495934 | 0.07 |
ENST00000218008.3 ENST00000361319.3 ENST00000539306.1 |
ATP1B4 |
ATPase, Na+/K+ transporting, beta 4 polypeptide |
| chr12_+_15475462 | 0.07 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chrX_-_49041242 | 0.07 |
ENST00000453382.1 ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
| chr1_+_161123536 | 0.07 |
ENST00000368003.5 |
UFC1 |
ubiquitin-fold modifier conjugating enzyme 1 |
| chrX_+_47004639 | 0.07 |
ENST00000345781.6 |
RBM10 |
RNA binding motif protein 10 |
| chr12_+_113344755 | 0.07 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr9_+_117350009 | 0.07 |
ENST00000374050.3 |
ATP6V1G1 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
| chrX_+_37850026 | 0.07 |
ENST00000341016.3 |
CXorf27 |
chromosome X open reading frame 27 |
| chrX_-_47004437 | 0.06 |
ENST00000276062.8 |
NDUFB11 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
| chr15_+_75628394 | 0.06 |
ENST00000564815.1 ENST00000338995.6 |
COMMD4 |
COMM domain containing 4 |
| chr12_-_48963829 | 0.06 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr15_+_75628232 | 0.06 |
ENST00000267935.8 ENST00000567195.1 |
COMMD4 |
COMM domain containing 4 |
| chr10_+_18429606 | 0.06 |
ENST00000324631.7 ENST00000352115.6 ENST00000377328.1 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_+_88529681 | 0.06 |
ENST00000399271.1 |
DSPP |
dentin sialophosphoprotein |
| chr11_+_71238313 | 0.06 |
ENST00000398536.4 |
KRTAP5-7 |
keratin associated protein 5-7 |
| chr7_+_141811539 | 0.05 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr1_+_22979474 | 0.05 |
ENST00000509305.1 |
C1QB |
complement component 1, q subcomponent, B chain |
| chr17_-_39041479 | 0.05 |
ENST00000167588.3 |
KRT20 |
keratin 20 |
| chr3_-_37034702 | 0.05 |
ENST00000322716.5 |
EPM2AIP1 |
EPM2A (laforin) interacting protein 1 |
| chr4_+_156588249 | 0.05 |
ENST00000393832.3 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr19_+_44576296 | 0.05 |
ENST00000421176.3 |
ZNF284 |
zinc finger protein 284 |
| chr6_+_25652501 | 0.05 |
ENST00000334979.6 |
SCGN |
secretagogin, EF-hand calcium binding protein |
| chr9_+_33629119 | 0.04 |
ENST00000331828.4 |
TRBV21OR9-2 |
T cell receptor beta variable 21/OR9-2 (pseudogene) |
| chr7_-_6048702 | 0.04 |
ENST00000265849.7 |
PMS2 |
PMS2 postmeiotic segregation increased 2 (S. cerevisiae) |
| chr2_-_39347524 | 0.04 |
ENST00000395038.2 ENST00000402219.2 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr4_+_156588115 | 0.04 |
ENST00000455639.2 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr17_+_41150290 | 0.04 |
ENST00000589037.1 ENST00000253788.5 |
RPL27 |
ribosomal protein L27 |
| chr5_-_179050066 | 0.04 |
ENST00000329433.6 ENST00000510411.1 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr5_+_54398463 | 0.03 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chrX_+_2984874 | 0.03 |
ENST00000359361.2 |
ARSF |
arylsulfatase F |
| chr6_+_42883727 | 0.03 |
ENST00000304672.1 ENST00000441198.1 ENST00000446507.1 |
PTCRA |
pre T-cell antigen receptor alpha |
| chr7_-_148725733 | 0.03 |
ENST00000286091.4 |
PDIA4 |
protein disulfide isomerase family A, member 4 |
| chr18_+_32455201 | 0.03 |
ENST00000590831.2 |
DTNA |
dystrobrevin, alpha |
| chr10_+_18429671 | 0.03 |
ENST00000282343.8 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr10_+_70715884 | 0.02 |
ENST00000354185.4 |
DDX21 |
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr13_-_99630233 | 0.02 |
ENST00000376460.1 ENST00000442173.1 |
DOCK9 |
dedicator of cytokinesis 9 |
| chrX_+_47004599 | 0.02 |
ENST00000329236.7 |
RBM10 |
RNA binding motif protein 10 |
| chr5_+_140800638 | 0.02 |
ENST00000398587.2 ENST00000518882.1 |
PCDHGA11 |
protocadherin gamma subfamily A, 11 |
| chr4_-_39367949 | 0.02 |
ENST00000503784.1 ENST00000349703.2 ENST00000381897.1 |
RFC1 |
replication factor C (activator 1) 1, 145kDa |
| chr15_+_67418047 | 0.02 |
ENST00000540846.2 |
SMAD3 |
SMAD family member 3 |
| chr6_+_29427548 | 0.02 |
ENST00000377132.1 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr8_-_21669826 | 0.02 |
ENST00000517328.1 |
GFRA2 |
GDNF family receptor alpha 2 |
| chr2_+_1418154 | 0.02 |
ENST00000423320.1 ENST00000382198.1 |
TPO |
thyroid peroxidase |
| chr17_+_68071389 | 0.02 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr9_-_128246769 | 0.01 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr19_+_41257084 | 0.01 |
ENST00000601393.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr3_+_37034823 | 0.01 |
ENST00000231790.2 ENST00000456676.2 |
MLH1 |
mutL homolog 1 |
| chr3_+_160394940 | 0.01 |
ENST00000320767.2 |
ARL14 |
ADP-ribosylation factor-like 14 |
| chr3_+_186435137 | 0.01 |
ENST00000447445.1 |
KNG1 |
kininogen 1 |
| chr5_+_135468516 | 0.01 |
ENST00000507118.1 ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5 |
SMAD family member 5 |
| chr2_-_49381572 | 0.01 |
ENST00000454032.1 ENST00000304421.4 |
FSHR |
follicle stimulating hormone receptor |
| chr13_-_45992541 | 0.01 |
ENST00000522438.1 |
SLC25A30 |
solute carrier family 25, member 30 |
| chr6_-_33385902 | 0.01 |
ENST00000374500.5 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr13_-_45992473 | 0.01 |
ENST00000539591.1 ENST00000519676.1 ENST00000519547.1 |
SLC25A30 |
solute carrier family 25, member 30 |
| chr19_+_4402659 | 0.01 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr19_-_57347415 | 0.01 |
ENST00000601070.1 |
ZIM2 |
zinc finger, imprinted 2 |
| chr3_-_184971853 | 0.01 |
ENST00000231887.3 |
EHHADH |
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr1_-_45988542 | 0.01 |
ENST00000424390.1 |
PRDX1 |
peroxiredoxin 1 |
| chr5_+_32711829 | 0.01 |
ENST00000415167.2 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr10_-_135187193 | 0.01 |
ENST00000368547.3 |
ECHS1 |
enoyl CoA hydratase, short chain, 1, mitochondrial |
| chr1_+_160051319 | 0.00 |
ENST00000368088.3 |
KCNJ9 |
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr8_+_24151620 | 0.00 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.2 | 2.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.7 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.1 | 4.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.1 | 0.6 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.6 | GO:0072143 | mesangial cell development(GO:0072143) glomerular mesangial cell development(GO:0072144) |
| 0.1 | 0.6 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.1 | 0.7 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.5 | GO:0098707 | ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.3 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 1.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.4 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.1 | 0.5 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:0090625 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 0.3 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 2.1 | GO:0050869 | negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.1 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.4 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.2 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.3 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 1.0 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.2 | GO:0060012 | glycine transport(GO:0015816) synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.3 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0090260 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.2 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.2 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.2 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.5 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.6 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.1 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.3 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.6 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 3.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.8 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.0 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.6 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.6 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.9 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 0.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 2.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 0.4 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.7 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.3 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.6 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 0.0 | 1.0 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.3 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.0 | 0.2 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 1.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.2 | 0.6 | GO:0004913 | interleukin-4 receptor activity(GO:0004913) interleukin-7 receptor activity(GO:0004917) |
| 0.2 | 0.5 | GO:0072510 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.2 | 0.7 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.4 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.1 | 0.4 | GO:0001016 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.1 | 0.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.6 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 0.2 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.2 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.4 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 2.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.1 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.0 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.3 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |