Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
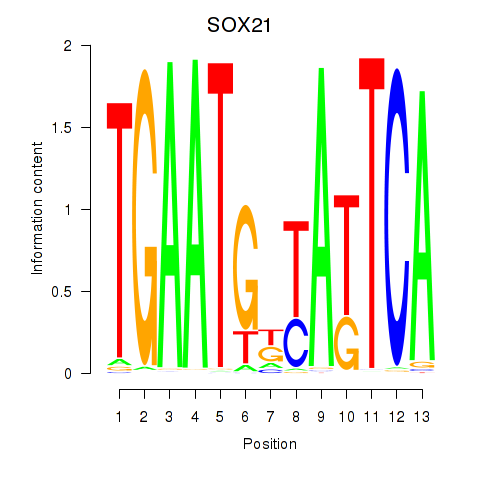
Results for SOX21
Z-value: 0.34
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.4 | SOX21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX21 | hg19_v2_chr13_-_95364389_95364389 | 0.00 | 1.0e+00 | Click! |
Activity profile of SOX21 motif
Sorted Z-values of SOX21 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX21
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_5005445 | 0.75 |
ENST00000380872.4 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
| chr6_-_112575912 | 0.56 |
ENST00000522006.1 ENST00000230538.7 ENST00000519932.1 |
LAMA4 |
laminin, alpha 4 |
| chr12_+_69742121 | 0.38 |
ENST00000261267.2 ENST00000549690.1 ENST00000548839.1 |
LYZ |
lysozyme |
| chr2_+_102759199 | 0.35 |
ENST00000409288.1 ENST00000410023.1 |
IL1R1 |
interleukin 1 receptor, type I |
| chr6_-_112575838 | 0.35 |
ENST00000455073.1 |
LAMA4 |
laminin, alpha 4 |
| chr8_-_19459993 | 0.29 |
ENST00000454498.2 ENST00000520003.1 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr20_+_43343886 | 0.29 |
ENST00000190983.4 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
| chr5_-_95158644 | 0.28 |
ENST00000237858.6 |
GLRX |
glutaredoxin (thioltransferase) |
| chr12_-_7261772 | 0.28 |
ENST00000545280.1 ENST00000543933.1 ENST00000545337.1 ENST00000544702.1 ENST00000266542.4 |
C1RL |
complement component 1, r subcomponent-like |
| chr6_-_112575758 | 0.26 |
ENST00000431543.2 ENST00000453937.2 ENST00000368638.4 ENST00000389463.4 |
LAMA4 |
laminin, alpha 4 |
| chrY_+_16634483 | 0.23 |
ENST00000382872.1 |
NLGN4Y |
neuroligin 4, Y-linked |
| chr7_+_120702819 | 0.23 |
ENST00000423795.1 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr1_+_221051699 | 0.22 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr5_-_95158375 | 0.21 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chr15_+_58702742 | 0.18 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr1_-_169455169 | 0.16 |
ENST00000367804.4 ENST00000236137.5 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
| chr7_-_86849883 | 0.15 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr3_-_155524049 | 0.14 |
ENST00000534941.1 ENST00000340171.2 |
C3orf33 |
chromosome 3 open reading frame 33 |
| chr6_+_131894284 | 0.13 |
ENST00000368087.3 ENST00000356962.2 |
ARG1 |
arginase 1 |
| chr3_-_37034702 | 0.13 |
ENST00000322716.5 |
EPM2AIP1 |
EPM2A (laforin) interacting protein 1 |
| chr12_-_10324716 | 0.12 |
ENST00000545927.1 ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1 |
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr1_+_146714291 | 0.12 |
ENST00000431239.1 ENST00000369259.3 ENST00000369258.4 ENST00000361293.5 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
| chr12_+_123011776 | 0.10 |
ENST00000450485.2 ENST00000333479.7 |
KNTC1 |
kinetochore associated 1 |
| chr5_-_55529115 | 0.10 |
ENST00000513241.2 ENST00000341048.4 |
ANKRD55 |
ankyrin repeat domain 55 |
| chr17_-_18266660 | 0.10 |
ENST00000582653.1 ENST00000352886.6 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr19_+_42381337 | 0.10 |
ENST00000597454.1 ENST00000444740.2 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr6_-_52926539 | 0.07 |
ENST00000350082.5 ENST00000356971.3 |
ICK |
intestinal cell (MAK-like) kinase |
| chr6_+_42531798 | 0.06 |
ENST00000372903.2 ENST00000372899.1 ENST00000372901.1 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr16_-_15149828 | 0.05 |
ENST00000566419.1 ENST00000568320.1 |
NTAN1 |
N-terminal asparagine amidase |
| chr19_+_42381173 | 0.05 |
ENST00000221972.3 |
CD79A |
CD79a molecule, immunoglobulin-associated alpha |
| chr8_+_67687413 | 0.04 |
ENST00000521960.1 ENST00000522398.1 ENST00000522629.1 ENST00000520976.1 ENST00000396596.1 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
| chr9_+_105757590 | 0.03 |
ENST00000374798.3 ENST00000487798.1 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr3_-_186288097 | 0.03 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr10_-_71169031 | 0.03 |
ENST00000373307.1 |
TACR2 |
tachykinin receptor 2 |
| chr15_-_80263506 | 0.03 |
ENST00000335661.6 |
BCL2A1 |
BCL2-related protein A1 |
| chr1_+_158901329 | 0.02 |
ENST00000368140.1 ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr11_-_8285405 | 0.02 |
ENST00000335790.3 ENST00000534484.1 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
| chr1_+_203764742 | 0.02 |
ENST00000432282.1 ENST00000453771.1 ENST00000367214.1 ENST00000367212.3 ENST00000332127.4 |
ZC3H11A |
zinc finger CCCH-type containing 11A |
| chr12_-_89746173 | 0.02 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr10_-_17243579 | 0.02 |
ENST00000525762.1 ENST00000412821.3 ENST00000351358.4 ENST00000377766.5 ENST00000358282.7 ENST00000488990.1 ENST00000377799.3 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
| chr6_+_31371337 | 0.02 |
ENST00000449934.2 ENST00000421350.1 |
MICA |
MHC class I polypeptide-related sequence A |
| chr3_-_164875850 | 0.01 |
ENST00000472120.1 |
RP11-747D18.1 |
RP11-747D18.1 |
| chr4_-_122854193 | 0.01 |
ENST00000513531.1 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr20_+_56964253 | 0.01 |
ENST00000395802.3 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr1_+_151171012 | 0.01 |
ENST00000349792.5 ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr4_+_81951957 | 0.01 |
ENST00000282701.2 |
BMP3 |
bone morphogenetic protein 3 |
| chrX_+_2984874 | 0.01 |
ENST00000359361.2 |
ARSF |
arylsulfatase F |
| chr14_-_107170409 | 0.01 |
ENST00000390633.2 |
IGHV1-69 |
immunoglobulin heavy variable 1-69 |
| chr2_+_3705785 | 0.00 |
ENST00000252505.3 |
ALLC |
allantoicase |
| chr1_+_67632083 | 0.00 |
ENST00000347310.5 ENST00000371002.1 |
IL23R |
interleukin 23 receptor |
| chrX_-_153210107 | 0.00 |
ENST00000369997.3 ENST00000393700.3 ENST00000412763.1 |
RENBP |
renin binding protein |
| chr12_-_1058685 | 0.00 |
ENST00000397230.2 ENST00000542785.1 ENST00000544742.1 ENST00000536177.1 ENST00000539046.1 ENST00000541619.1 |
RAD52 |
RAD52 homolog (S. cerevisiae) |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0047023 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.1 | 0.3 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.1 | 0.4 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.1 | 0.4 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.2 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.5 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.1 | 0.8 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.1 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.1 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.5 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 0.0 | 0.2 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.1 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.0 | 0.2 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.0 | GO:0033150 | perinuclear theca(GO:0033011) cytoskeletal calyx(GO:0033150) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |