Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
Results for SOX30
Z-value: 0.68
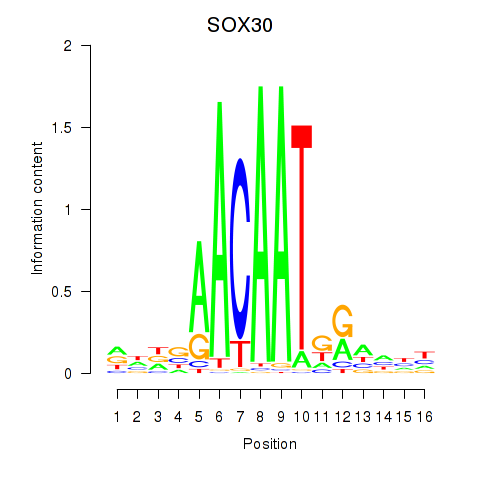
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.6 | SOX30 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX30 | hg19_v2_chr5_-_157079372_157079395, hg19_v2_chr5_-_157079428_157079452 | 0.34 | 1.9e-01 | Click! |
Activity profile of SOX30 motif
Sorted Z-values of SOX30 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX30
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_12290586 | 1.66 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr11_-_89224638 | 1.40 |
ENST00000535633.1 ENST00000263317.4 |
NOX4 |
NADPH oxidase 4 |
| chr11_+_46402744 | 1.01 |
ENST00000533952.1 |
MDK |
midkine (neurite growth-promoting factor 2) |
| chr12_-_10251603 | 0.86 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr11_+_128563948 | 0.86 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr11_+_128563652 | 0.85 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr12_-_10251576 | 0.83 |
ENST00000315330.4 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr5_-_141338627 | 0.81 |
ENST00000231484.3 |
PCDH12 |
protocadherin 12 |
| chr7_-_11871815 | 0.66 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr12_-_10251539 | 0.61 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr17_+_67498538 | 0.54 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr11_-_75236867 | 0.53 |
ENST00000376282.3 ENST00000336898.3 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr20_-_45981138 | 0.44 |
ENST00000446994.2 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr11_-_102323489 | 0.40 |
ENST00000361236.3 |
TMEM123 |
transmembrane protein 123 |
| chr11_-_102323740 | 0.37 |
ENST00000398136.2 |
TMEM123 |
transmembrane protein 123 |
| chr11_+_125365110 | 0.35 |
ENST00000527818.1 |
AP000708.1 |
AP000708.1 |
| chr1_+_244998602 | 0.35 |
ENST00000411948.2 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
| chr17_+_47448102 | 0.34 |
ENST00000576461.1 |
RP11-81K2.1 |
Uncharacterized protein |
| chr4_+_113152978 | 0.33 |
ENST00000309703.6 |
AP1AR |
adaptor-related protein complex 1 associated regulatory protein |
| chr1_+_93913713 | 0.33 |
ENST00000604705.1 ENST00000370253.2 |
FNBP1L |
formin binding protein 1-like |
| chrY_+_15016013 | 0.32 |
ENST00000360160.4 ENST00000454054.1 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr8_-_141810634 | 0.31 |
ENST00000521986.1 ENST00000523539.1 ENST00000538769.1 |
PTK2 |
protein tyrosine kinase 2 |
| chrX_-_15872914 | 0.31 |
ENST00000380291.1 ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_+_143635067 | 0.25 |
ENST00000264170.4 |
KYNU |
kynureninase |
| chr3_-_8686479 | 0.25 |
ENST00000544814.1 ENST00000427408.1 |
SSUH2 |
ssu-2 homolog (C. elegans) |
| chr2_+_232573222 | 0.24 |
ENST00000341369.7 ENST00000409683.1 |
PTMA |
prothymosin, alpha |
| chr2_+_232573208 | 0.23 |
ENST00000409115.3 |
PTMA |
prothymosin, alpha |
| chr2_-_169769787 | 0.21 |
ENST00000451987.1 |
SPC25 |
SPC25, NDC80 kinetochore complex component |
| chr10_+_35484793 | 0.21 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr9_+_103235365 | 0.20 |
ENST00000374879.4 |
TMEFF1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_+_74701062 | 0.20 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr10_+_70480963 | 0.19 |
ENST00000265872.6 ENST00000535016.1 ENST00000538031.1 ENST00000543719.1 ENST00000539539.1 ENST00000543225.1 ENST00000536012.1 ENST00000494903.2 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
| chr2_+_86669118 | 0.17 |
ENST00000427678.1 ENST00000542128.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr8_-_82024290 | 0.17 |
ENST00000220597.4 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
| chr6_+_22146851 | 0.16 |
ENST00000606197.1 |
CASC15 |
cancer susceptibility candidate 15 (non-protein coding) |
| chr13_-_41593425 | 0.15 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr1_-_243418344 | 0.15 |
ENST00000366542.1 |
CEP170 |
centrosomal protein 170kDa |
| chr22_-_43485381 | 0.15 |
ENST00000331018.7 ENST00000266254.7 ENST00000445824.1 |
TTLL1 |
tubulin tyrosine ligase-like family, member 1 |
| chr7_+_100770328 | 0.15 |
ENST00000223095.4 ENST00000445463.2 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr22_-_30970560 | 0.13 |
ENST00000402369.1 ENST00000406361.1 |
GAL3ST1 |
galactose-3-O-sulfotransferase 1 |
| chr6_-_89927151 | 0.13 |
ENST00000454853.2 |
GABRR1 |
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr1_+_185014496 | 0.12 |
ENST00000367510.3 |
RNF2 |
ring finger protein 2 |
| chr1_-_243418621 | 0.12 |
ENST00000366544.1 ENST00000366543.1 |
CEP170 |
centrosomal protein 170kDa |
| chr10_+_35416090 | 0.12 |
ENST00000354759.3 |
CREM |
cAMP responsive element modulator |
| chr1_-_156828810 | 0.11 |
ENST00000368195.3 |
INSRR |
insulin receptor-related receptor |
| chr10_+_35415719 | 0.11 |
ENST00000474362.1 ENST00000374721.3 |
CREM |
cAMP responsive element modulator |
| chr5_+_66254698 | 0.10 |
ENST00000405643.1 ENST00000407621.1 ENST00000432426.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr2_-_157198860 | 0.10 |
ENST00000409572.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr4_-_70725856 | 0.09 |
ENST00000226444.3 |
SULT1E1 |
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr4_-_87028478 | 0.08 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr17_-_10372875 | 0.06 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr16_+_87636474 | 0.06 |
ENST00000284262.2 |
JPH3 |
junctophilin 3 |
| chr5_-_160279207 | 0.06 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chr1_+_180897269 | 0.06 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chrX_+_41192595 | 0.06 |
ENST00000399959.2 |
DDX3X |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, X-linked |
| chr10_+_35415978 | 0.05 |
ENST00000429130.3 ENST00000469949.2 ENST00000460270.1 |
CREM |
cAMP responsive element modulator |
| chr5_+_170288856 | 0.05 |
ENST00000523189.1 |
RANBP17 |
RAN binding protein 17 |
| chr11_-_102576537 | 0.05 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr12_-_10282742 | 0.05 |
ENST00000298523.5 ENST00000396484.2 ENST00000310002.4 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr10_+_35416223 | 0.05 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr12_-_11422739 | 0.05 |
ENST00000279573.7 |
PRB3 |
proline-rich protein BstNI subfamily 3 |
| chr12_-_11036844 | 0.04 |
ENST00000428168.2 |
PRH1 |
proline-rich protein HaeIII subfamily 1 |
| chr1_-_167883327 | 0.04 |
ENST00000476818.2 ENST00000367851.4 ENST00000367848.1 |
ADCY10 |
adenylate cyclase 10 (soluble) |
| chr14_-_90421028 | 0.04 |
ENST00000267544.9 ENST00000316738.7 ENST00000538485.2 ENST00000556609.1 |
EFCAB11 |
EF-hand calcium binding domain 11 |
| chr1_+_228337553 | 0.03 |
ENST00000366714.2 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
| chr18_+_32455201 | 0.03 |
ENST00000590831.2 |
DTNA |
dystrobrevin, alpha |
| chr11_+_63137251 | 0.03 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr14_+_101299520 | 0.02 |
ENST00000455531.1 |
MEG3 |
maternally expressed 3 (non-protein coding) |
| chr2_+_27719697 | 0.02 |
ENST00000264717.2 ENST00000424318.2 |
GCKR |
glucokinase (hexokinase 4) regulator |
| chr3_-_167191814 | 0.01 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr14_+_52456327 | 0.01 |
ENST00000556760.1 |
C14orf166 |
chromosome 14 open reading frame 166 |
| chr6_+_126240442 | 0.00 |
ENST00000448104.1 ENST00000438495.1 ENST00000444128.1 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr14_-_90420862 | 0.00 |
ENST00000556005.1 ENST00000555872.1 |
EFCAB11 |
EF-hand calcium binding domain 11 |
| chr12_-_11422630 | 0.00 |
ENST00000381842.3 ENST00000538488.1 |
PRB3 |
proline-rich protein BstNI subfamily 3 |
| chr1_+_154401791 | 0.00 |
ENST00000476006.1 |
IL6R |
interleukin 6 receptor |
Gene Ontology Analysis
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 1.4 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.1 | 0.5 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.3 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.3 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0071535 | RING-like zinc finger domain binding(GO:0071535) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 0.5 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.2 | 1.0 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.3 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.3 | GO:0097052 | tryptophan catabolic process to acetyl-CoA(GO:0019442) L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 1.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 0.4 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:2000097 | chronological cell aging(GO:0001300) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.0 | 0.5 | GO:0048505 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 1.4 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0021986 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.1 | 1.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.5 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |