Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
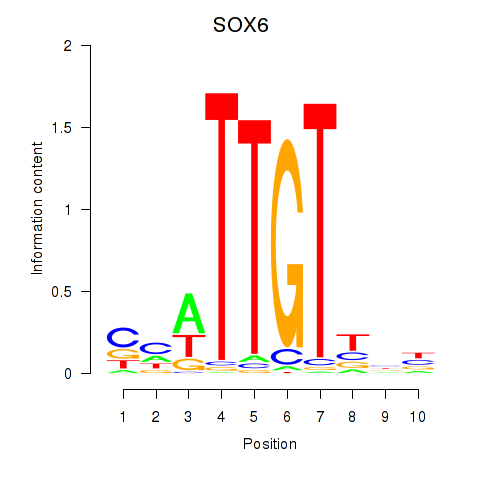
Results for SOX6
Z-value: 1.44
Transcription factors associated with SOX6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX6
|
ENSG00000110693.11 | SOX6 |
Activity profile of SOX6 motif
Sorted Z-values of SOX6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_37899323 | 4.02 |
ENST00000295324.3 ENST00000457889.1 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
| chrX_+_12993202 | 3.03 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chrX_+_65384052 | 2.97 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chrX_+_65384182 | 2.91 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr9_+_139874683 | 2.63 |
ENST00000444903.1 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr6_-_24877490 | 2.25 |
ENST00000540914.1 ENST00000378023.4 |
FAM65B |
family with sequence similarity 65, member B |
| chrX_+_12993336 | 2.10 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr12_-_8815215 | 2.05 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr12_-_8815299 | 2.04 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr6_+_13272904 | 1.94 |
ENST00000379335.3 ENST00000379329.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr7_+_69064300 | 1.88 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr2_-_86564776 | 1.86 |
ENST00000165698.5 ENST00000541910.1 ENST00000535845.1 |
REEP1 |
receptor accessory protein 1 |
| chr2_-_9143786 | 1.86 |
ENST00000462696.1 ENST00000305997.3 |
MBOAT2 |
membrane bound O-acyltransferase domain containing 2 |
| chr11_-_10830463 | 1.85 |
ENST00000527419.1 ENST00000530211.1 ENST00000530702.1 ENST00000524932.1 ENST00000532570.1 |
EIF4G2 |
eukaryotic translation initiation factor 4 gamma, 2 |
| chr5_-_157002775 | 1.77 |
ENST00000257527.4 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr12_-_8815404 | 1.48 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr15_+_80696666 | 1.46 |
ENST00000303329.4 |
ARNT2 |
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr17_+_52978156 | 1.43 |
ENST00000348161.4 |
TOM1L1 |
target of myb1 (chicken)-like 1 |
| chr4_+_41614909 | 1.37 |
ENST00000509454.1 ENST00000396595.3 ENST00000381753.4 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr3_-_114790179 | 1.20 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr7_+_23146271 | 1.14 |
ENST00000545771.1 |
KLHL7 |
kelch-like family member 7 |
| chr15_+_63335899 | 1.14 |
ENST00000561266.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr9_+_101705893 | 1.13 |
ENST00000375001.3 |
COL15A1 |
collagen, type XV, alpha 1 |
| chr12_+_104359641 | 1.09 |
ENST00000537100.1 |
TDG |
thymine-DNA glycosylase |
| chr7_+_102553430 | 1.09 |
ENST00000339431.4 ENST00000249377.4 |
LRRC17 |
leucine rich repeat containing 17 |
| chr14_-_71107921 | 1.08 |
ENST00000553982.1 ENST00000500016.1 |
CTD-2540L5.5 CTD-2540L5.6 |
CTD-2540L5.5 CTD-2540L5.6 |
| chr18_+_6729698 | 1.03 |
ENST00000383472.4 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr2_-_217560248 | 1.03 |
ENST00000233813.4 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
| chr4_-_109090106 | 1.03 |
ENST00000379951.2 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr1_+_182808474 | 0.98 |
ENST00000367549.3 |
DHX9 |
DEAH (Asp-Glu-Ala-His) box helicase 9 |
| chr4_-_143226979 | 0.94 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr5_-_157002749 | 0.93 |
ENST00000517905.1 ENST00000430702.2 ENST00000394020.1 |
ADAM19 |
ADAM metallopeptidase domain 19 |
| chr12_+_69979446 | 0.91 |
ENST00000543146.2 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
| chr4_-_109089573 | 0.88 |
ENST00000265165.1 |
LEF1 |
lymphoid enhancer-binding factor 1 |
| chr4_-_103266355 | 0.87 |
ENST00000424970.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr10_+_22605374 | 0.86 |
ENST00000448361.1 |
COMMD3 |
COMM domain containing 3 |
| chr12_+_104359576 | 0.86 |
ENST00000392872.3 ENST00000436021.2 |
TDG |
thymine-DNA glycosylase |
| chr21_+_30671690 | 0.85 |
ENST00000399921.1 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr5_-_169725231 | 0.84 |
ENST00000046794.5 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
| chr12_+_104359614 | 0.84 |
ENST00000266775.9 ENST00000544861.1 |
TDG |
thymine-DNA glycosylase |
| chr11_+_131781290 | 0.82 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr1_+_14075903 | 0.80 |
ENST00000343137.4 ENST00000503842.1 ENST00000407521.3 ENST00000505823.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chr4_+_26585538 | 0.80 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr4_-_143227088 | 0.79 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr16_-_67970990 | 0.79 |
ENST00000358514.4 |
PSMB10 |
proteasome (prosome, macropain) subunit, beta type, 10 |
| chr3_+_133118839 | 0.79 |
ENST00000302334.2 |
BFSP2 |
beaded filament structural protein 2, phakinin |
| chr11_-_79151695 | 0.78 |
ENST00000278550.7 |
TENM4 |
teneurin transmembrane protein 4 |
| chr4_-_105416039 | 0.77 |
ENST00000394767.2 |
CXXC4 |
CXXC finger protein 4 |
| chr4_+_26585686 | 0.77 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr15_+_89631647 | 0.76 |
ENST00000569550.1 ENST00000565066.1 ENST00000565973.1 |
ABHD2 |
abhydrolase domain containing 2 |
| chr12_-_49582593 | 0.72 |
ENST00000295766.5 |
TUBA1A |
tubulin, alpha 1a |
| chrX_+_129473916 | 0.69 |
ENST00000545805.1 ENST00000543953.1 ENST00000218197.5 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr14_+_103589789 | 0.69 |
ENST00000558056.1 ENST00000560869.1 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
| chr15_+_79166065 | 0.68 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr16_+_56970567 | 0.68 |
ENST00000563911.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr5_+_49962772 | 0.67 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr10_+_22605304 | 0.65 |
ENST00000475460.2 ENST00000602390.1 ENST00000489125.2 ENST00000456711.1 ENST00000444869.1 |
COMMD3-BMI1 COMMD3 |
COMMD3-BMI1 readthrough COMM domain containing 3 |
| chr10_+_35416223 | 0.65 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chr4_-_103266219 | 0.65 |
ENST00000394833.2 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr14_-_51027838 | 0.64 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr17_+_30813576 | 0.63 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr1_+_14075865 | 0.62 |
ENST00000413440.1 |
PRDM2 |
PR domain containing 2, with ZNF domain |
| chr13_-_24007815 | 0.62 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr14_-_106926724 | 0.62 |
ENST00000434710.1 |
IGHV3-43 |
immunoglobulin heavy variable 3-43 |
| chr9_+_108463234 | 0.62 |
ENST00000374688.1 |
TMEM38B |
transmembrane protein 38B |
| chrX_+_135579670 | 0.62 |
ENST00000218364.4 |
HTATSF1 |
HIV-1 Tat specific factor 1 |
| chr7_-_150924121 | 0.62 |
ENST00000441774.1 ENST00000222388.2 ENST00000287844.2 |
ABCF2 |
ATP-binding cassette, sub-family F (GCN20), member 2 |
| chr1_-_26232951 | 0.59 |
ENST00000426559.2 ENST00000455785.2 |
STMN1 |
stathmin 1 |
| chr16_+_33605231 | 0.57 |
ENST00000570121.2 |
IGHV3OR16-12 |
immunoglobulin heavy variable 3/OR16-12 (non-functional) |
| chr1_-_154155675 | 0.57 |
ENST00000330188.9 ENST00000341485.5 |
TPM3 |
tropomyosin 3 |
| chr12_-_49582978 | 0.56 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chrX_-_106959631 | 0.56 |
ENST00000486554.1 ENST00000372390.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr8_-_120685608 | 0.56 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr1_+_181003067 | 0.56 |
ENST00000434571.2 ENST00000367579.3 ENST00000282990.6 ENST00000367580.5 |
MR1 |
major histocompatibility complex, class I-related |
| chr5_-_114515734 | 0.56 |
ENST00000514154.1 ENST00000282369.3 |
TRIM36 |
tripartite motif containing 36 |
| chr15_+_52311398 | 0.55 |
ENST00000261845.5 |
MAPK6 |
mitogen-activated protein kinase 6 |
| chr19_-_18717627 | 0.55 |
ENST00000392386.3 |
CRLF1 |
cytokine receptor-like factor 1 |
| chr1_+_150039369 | 0.54 |
ENST00000369130.3 ENST00000369128.5 |
VPS45 |
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr5_-_76788317 | 0.54 |
ENST00000296679.4 |
WDR41 |
WD repeat domain 41 |
| chr4_+_54243798 | 0.52 |
ENST00000337488.6 ENST00000358575.5 ENST00000507922.1 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
| chrX_+_129473859 | 0.52 |
ENST00000424447.1 |
SLC25A14 |
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chrX_-_101726732 | 0.52 |
ENST00000457521.2 ENST00000412230.2 ENST00000453326.2 |
NXF2B TCP11X2 |
nuclear RNA export factor 2B t-complex 11 family, X-linked 2 |
| chr11_+_125496619 | 0.52 |
ENST00000532669.1 ENST00000278916.3 |
CHEK1 |
checkpoint kinase 1 |
| chr13_-_41593425 | 0.52 |
ENST00000239882.3 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
| chr14_-_106518922 | 0.51 |
ENST00000390598.2 |
IGHV3-7 |
immunoglobulin heavy variable 3-7 |
| chr18_+_6729725 | 0.51 |
ENST00000400091.2 ENST00000583410.1 ENST00000584387.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chrX_+_80457442 | 0.50 |
ENST00000373212.5 |
SH3BGRL |
SH3 domain binding glutamic acid-rich protein like |
| chrX_+_135579238 | 0.50 |
ENST00000535601.1 ENST00000448450.1 ENST00000425695.1 |
HTATSF1 |
HIV-1 Tat specific factor 1 |
| chrX_+_101470280 | 0.49 |
ENST00000395088.2 ENST00000330252.5 ENST00000333110.5 |
NXF2 TCP11X1 |
nuclear RNA export factor 2 t-complex 11 family, X-linked 1 |
| chr12_+_25205666 | 0.49 |
ENST00000547044.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr4_+_54243862 | 0.49 |
ENST00000306932.6 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
| chr11_-_64013663 | 0.48 |
ENST00000392210.2 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr6_+_122720681 | 0.48 |
ENST00000368455.4 ENST00000452194.1 |
HSF2 |
heat shock transcription factor 2 |
| chr5_-_146435694 | 0.48 |
ENST00000356826.3 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr5_-_176836577 | 0.48 |
ENST00000253496.3 |
F12 |
coagulation factor XII (Hageman factor) |
| chr20_-_3996036 | 0.47 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chr4_+_71570430 | 0.47 |
ENST00000417478.2 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr16_-_30798492 | 0.47 |
ENST00000262525.4 |
ZNF629 |
zinc finger protein 629 |
| chr1_-_32801825 | 0.47 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr15_+_89631381 | 0.45 |
ENST00000352732.5 |
ABHD2 |
abhydrolase domain containing 2 |
| chr1_-_155880672 | 0.45 |
ENST00000609492.1 ENST00000368322.3 |
RIT1 |
Ras-like without CAAX 1 |
| chr4_-_71705060 | 0.45 |
ENST00000514161.1 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr21_-_40720974 | 0.44 |
ENST00000380748.1 |
HMGN1 |
high mobility group nucleosome binding domain 1 |
| chr4_-_71705027 | 0.44 |
ENST00000545193.1 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
| chr16_+_54964740 | 0.42 |
ENST00000394636.4 |
IRX5 |
iroquois homeobox 5 |
| chr6_-_109777128 | 0.42 |
ENST00000358807.3 ENST00000358577.3 |
MICAL1 |
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr1_+_25071848 | 0.41 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr6_-_133055815 | 0.41 |
ENST00000509351.1 ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3 |
vanin 3 |
| chr8_+_42195972 | 0.40 |
ENST00000532157.1 ENST00000265421.4 ENST00000520008.1 |
POLB |
polymerase (DNA directed), beta |
| chr14_-_106845789 | 0.40 |
ENST00000390617.2 |
IGHV3-35 |
immunoglobulin heavy variable 3-35 (non-functional) |
| chr9_-_88896977 | 0.40 |
ENST00000311534.6 |
ISCA1 |
iron-sulfur cluster assembly 1 |
| chr8_+_24151620 | 0.38 |
ENST00000437154.2 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr4_+_71588372 | 0.38 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr14_-_106725723 | 0.37 |
ENST00000390609.2 |
IGHV3-23 |
immunoglobulin heavy variable 3-23 |
| chr17_+_75283973 | 0.37 |
ENST00000431235.2 ENST00000449803.2 |
SEPT9 |
septin 9 |
| chr3_+_181429704 | 0.37 |
ENST00000431565.2 ENST00000325404.1 |
SOX2 |
SRY (sex determining region Y)-box 2 |
| chr7_+_99006232 | 0.37 |
ENST00000403633.2 |
BUD31 |
BUD31 homolog (S. cerevisiae) |
| chr7_-_99699538 | 0.36 |
ENST00000343023.6 ENST00000303887.5 |
MCM7 |
minichromosome maintenance complex component 7 |
| chr12_+_15475462 | 0.35 |
ENST00000543886.1 ENST00000348962.2 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr14_-_91884150 | 0.35 |
ENST00000553403.1 |
CCDC88C |
coiled-coil domain containing 88C |
| chr1_+_24286287 | 0.34 |
ENST00000334351.7 ENST00000374468.1 |
PNRC2 |
proline-rich nuclear receptor coactivator 2 |
| chr1_-_89357179 | 0.34 |
ENST00000448623.1 ENST00000418217.1 ENST00000370500.5 |
GTF2B |
general transcription factor IIB |
| chr22_+_43808014 | 0.33 |
ENST00000334209.5 ENST00000443721.1 ENST00000414469.2 ENST00000439548.1 |
MPPED1 |
metallophosphoesterase domain containing 1 |
| chr4_+_78078304 | 0.33 |
ENST00000316355.5 ENST00000354403.5 ENST00000502280.1 |
CCNG2 |
cyclin G2 |
| chr21_-_16437126 | 0.32 |
ENST00000318948.4 |
NRIP1 |
nuclear receptor interacting protein 1 |
| chr2_-_157198860 | 0.32 |
ENST00000409572.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr7_+_77167343 | 0.32 |
ENST00000433369.2 ENST00000415482.2 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr10_-_62332357 | 0.31 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_-_197024394 | 0.31 |
ENST00000434148.1 ENST00000412364.2 ENST00000436682.1 ENST00000456699.2 ENST00000392380.2 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr16_+_15528332 | 0.31 |
ENST00000566490.1 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr9_-_130639997 | 0.31 |
ENST00000373176.1 |
AK1 |
adenylate kinase 1 |
| chr8_-_20161466 | 0.31 |
ENST00000381569.1 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
| chr11_-_102576537 | 0.29 |
ENST00000260229.4 |
MMP27 |
matrix metallopeptidase 27 |
| chr12_-_11508520 | 0.28 |
ENST00000545626.1 ENST00000500254.2 |
PRB1 |
proline-rich protein BstNI subfamily 1 |
| chr21_-_40720995 | 0.28 |
ENST00000380749.5 |
HMGN1 |
high mobility group nucleosome binding domain 1 |
| chr8_+_24151553 | 0.28 |
ENST00000265769.4 ENST00000540823.1 ENST00000397649.3 |
ADAM28 |
ADAM metallopeptidase domain 28 |
| chr2_+_168725458 | 0.28 |
ENST00000392690.3 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr21_-_16437255 | 0.28 |
ENST00000400199.1 ENST00000400202.1 |
NRIP1 |
nuclear receptor interacting protein 1 |
| chr17_+_75447326 | 0.28 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chr12_+_69633372 | 0.28 |
ENST00000456847.3 ENST00000266679.8 |
CPSF6 |
cleavage and polyadenylation specific factor 6, 68kDa |
| chr12_-_105478339 | 0.28 |
ENST00000424857.2 ENST00000258494.9 |
ALDH1L2 |
aldehyde dehydrogenase 1 family, member L2 |
| chr16_+_89989687 | 0.28 |
ENST00000315491.7 ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3 |
Tubulin beta-3 chain |
| chr7_+_107224364 | 0.28 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr12_-_58146048 | 0.28 |
ENST00000547281.1 ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4 |
cyclin-dependent kinase 4 |
| chr6_-_3227877 | 0.27 |
ENST00000259818.7 |
TUBB2B |
tubulin, beta 2B class IIb |
| chrX_+_84498989 | 0.27 |
ENST00000395402.1 |
ZNF711 |
zinc finger protein 711 |
| chr3_+_151986709 | 0.27 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr22_-_42336209 | 0.26 |
ENST00000472374.2 |
CENPM |
centromere protein M |
| chr2_+_170590321 | 0.26 |
ENST00000392647.2 |
KLHL23 |
kelch-like family member 23 |
| chr17_+_75372165 | 0.26 |
ENST00000427674.2 |
SEPT9 |
septin 9 |
| chr19_-_51891209 | 0.26 |
ENST00000221973.3 ENST00000596399.1 |
LIM2 |
lens intrinsic membrane protein 2, 19kDa |
| chr5_-_146435572 | 0.25 |
ENST00000394414.1 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr15_+_85923856 | 0.25 |
ENST00000560302.1 ENST00000394518.2 ENST00000361243.2 ENST00000560256.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr12_+_69633407 | 0.25 |
ENST00000551516.1 |
CPSF6 |
cleavage and polyadenylation specific factor 6, 68kDa |
| chr17_+_9066252 | 0.25 |
ENST00000436734.1 |
NTN1 |
netrin 1 |
| chr5_-_146435501 | 0.25 |
ENST00000336640.6 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
| chr17_-_40288449 | 0.24 |
ENST00000552162.1 ENST00000550504.1 |
RAB5C |
RAB5C, member RAS oncogene family |
| chr15_+_64388166 | 0.24 |
ENST00000353874.4 ENST00000261889.5 ENST00000559844.1 ENST00000561026.1 ENST00000558040.1 |
SNX1 |
sorting nexin 1 |
| chr2_-_47572105 | 0.23 |
ENST00000419035.1 ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4 |
AC073283.4 |
| chr1_-_26324534 | 0.23 |
ENST00000374284.1 ENST00000441420.1 ENST00000374282.3 |
PAFAH2 |
platelet-activating factor acetylhydrolase 2, 40kDa |
| chr14_-_106866934 | 0.22 |
ENST00000390618.2 |
IGHV3-38 |
immunoglobulin heavy variable 3-38 (non-functional) |
| chr1_-_26232522 | 0.22 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr16_-_29478016 | 0.22 |
ENST00000549858.1 ENST00000551411.1 |
RP11-345J4.3 |
Uncharacterized protein |
| chr5_+_140213815 | 0.22 |
ENST00000525929.1 ENST00000378125.3 |
PCDHA7 |
protocadherin alpha 7 |
| chr1_-_212004090 | 0.22 |
ENST00000366997.4 |
LPGAT1 |
lysophosphatidylglycerol acyltransferase 1 |
| chr11_+_63706444 | 0.21 |
ENST00000377793.4 ENST00000456907.2 ENST00000539656.1 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
| chr17_+_21729593 | 0.21 |
ENST00000581769.1 ENST00000584755.1 |
UBBP4 |
ubiquitin B pseudogene 4 |
| chr16_-_3068171 | 0.20 |
ENST00000572154.1 ENST00000328796.4 |
CLDN6 |
claudin 6 |
| chr4_+_144303093 | 0.19 |
ENST00000505913.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr5_-_39425068 | 0.19 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr6_-_31782813 | 0.19 |
ENST00000375654.4 |
HSPA1L |
heat shock 70kDa protein 1-like |
| chr3_-_123339418 | 0.19 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr17_-_49124230 | 0.19 |
ENST00000510283.1 ENST00000510855.1 |
SPAG9 |
sperm associated antigen 9 |
| chr6_-_49712123 | 0.19 |
ENST00000263045.4 |
CRISP3 |
cysteine-rich secretory protein 3 |
| chr12_-_99288536 | 0.19 |
ENST00000549797.1 ENST00000333732.7 ENST00000341752.7 |
ANKS1B |
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr4_+_70861647 | 0.19 |
ENST00000246895.4 ENST00000381060.2 |
STATH |
statherin |
| chr1_+_154955797 | 0.19 |
ENST00000368433.1 ENST00000315144.10 ENST00000368432.1 ENST00000368431.3 ENST00000292180.3 |
FLAD1 |
flavin adenine dinucleotide synthetase 1 |
| chr12_-_11548496 | 0.18 |
ENST00000389362.4 ENST00000565533.1 ENST00000546254.1 |
PRB2 PRB1 |
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr7_+_87563557 | 0.18 |
ENST00000439864.1 ENST00000412441.1 ENST00000398201.4 ENST00000265727.7 ENST00000315984.7 ENST00000398209.3 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr19_+_5681153 | 0.18 |
ENST00000579559.1 ENST00000577222.1 |
HSD11B1L RPL36 |
hydroxysteroid (11-beta) dehydrogenase 1-like ribosomal protein L36 |
| chr3_-_123339343 | 0.18 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr6_-_135375921 | 0.17 |
ENST00000367820.2 ENST00000314674.3 ENST00000524715.1 ENST00000415177.2 ENST00000367826.2 |
HBS1L |
HBS1-like (S. cerevisiae) |
| chr6_-_127840453 | 0.17 |
ENST00000556132.1 |
SOGA3 |
SOGA family member 3 |
| chr11_+_65339820 | 0.17 |
ENST00000316409.2 ENST00000449319.2 ENST00000530349.1 |
FAM89B |
family with sequence similarity 89, member B |
| chr16_-_29466285 | 0.17 |
ENST00000330978.3 |
BOLA2 |
bolA family member 2 |
| chr9_-_123476719 | 0.17 |
ENST00000373930.3 |
MEGF9 |
multiple EGF-like-domains 9 |
| chr1_+_196743912 | 0.17 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr5_+_134094461 | 0.17 |
ENST00000452510.2 ENST00000354283.4 |
DDX46 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr20_+_20348740 | 0.16 |
ENST00000310227.1 |
INSM1 |
insulinoma-associated 1 |
| chr7_+_87563458 | 0.16 |
ENST00000398204.4 |
ADAM22 |
ADAM metallopeptidase domain 22 |
| chr8_-_30515693 | 0.16 |
ENST00000355904.4 |
GTF2E2 |
general transcription factor IIE, polypeptide 2, beta 34kDa |
| chr4_+_41614720 | 0.16 |
ENST00000509277.1 |
LIMCH1 |
LIM and calponin homology domains 1 |
| chr17_-_62308087 | 0.15 |
ENST00000583097.1 |
TEX2 |
testis expressed 2 |
| chr19_+_35783047 | 0.15 |
ENST00000595791.1 ENST00000597035.1 ENST00000537831.2 |
MAG |
myelin associated glycoprotein |
| chr2_+_70056762 | 0.15 |
ENST00000282570.3 |
GMCL1 |
germ cell-less, spermatogenesis associated 1 |
| chr19_+_35783037 | 0.15 |
ENST00000361922.4 |
MAG |
myelin associated glycoprotein |
| chrX_+_70503433 | 0.14 |
ENST00000276079.8 ENST00000373856.3 ENST00000373841.1 ENST00000420903.1 |
NONO |
non-POU domain containing, octamer-binding |
| chr22_+_32439019 | 0.14 |
ENST00000266088.4 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr6_-_89927151 | 0.14 |
ENST00000454853.2 |
GABRR1 |
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chrX_-_153599578 | 0.14 |
ENST00000360319.4 ENST00000344736.4 |
FLNA |
filamin A, alpha |
Gene Ontology Analysis
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.6 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 3.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.7 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.9 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 5.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.7 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.0 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.2 | 1.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 0.5 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.8 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 0.6 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.4 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.1 | 1.0 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.7 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.5 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.9 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 0.9 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.6 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 1.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.1 | 0.7 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 1.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.1 | 0.6 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 1.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 5.1 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 2.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.3 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.8 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 1.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 2.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 4.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0035577 | azurophil granule membrane(GO:0035577) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0005876 | spindle microtubule(GO:0005876) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 2.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.3 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 4.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.7 | 2.6 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.5 | 4.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.3 | 1.7 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.3 | 2.1 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.2 | 4.0 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.2 | 0.6 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.2 | 1.0 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.1 | 0.6 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.5 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 1.9 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 5.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.5 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.5 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 1.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.1 | 0.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.2 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.6 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.5 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 2.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 3.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.3 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.9 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 2.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.1 | GO:0036134 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.3 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.3 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.5 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.7 | 2.8 | GO:1902544 | oxidative DNA demethylation(GO:0035511) regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.5 | 1.5 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.5 | 1.9 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.5 | 1.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 1.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.3 | 0.9 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.3 | 3.6 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.3 | 0.8 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.3 | 1.5 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 1.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.2 | 0.6 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.2 | 1.0 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.2 | 0.5 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.2 | 4.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 2.7 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 1.1 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 5.6 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.1 | 0.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.5 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.4 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.1 | 0.8 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 1.9 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.4 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.2 | GO:0036148 | phosphatidylglycerol acyl-chain remodeling(GO:0036148) |
| 0.1 | 0.3 | GO:0021538 | epithalamus development(GO:0021538) habenula development(GO:0021986) |
| 0.1 | 0.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.6 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.1 | 1.0 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 1.4 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 0.4 | GO:1904636 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.3 | GO:0072658 | positive regulation of cell communication by electrical coupling(GO:0010650) maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.6 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.1 | 1.4 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 1.9 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.1 | 0.2 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.2 | GO:0042727 | flavin-containing compound biosynthetic process(GO:0042727) |
| 0.1 | 0.7 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.3 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.4 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.9 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.1 | 0.4 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.1 | 0.3 | GO:1904798 | positive regulation of core promoter binding(GO:1904798) |
| 0.1 | 2.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.3 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.7 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 1.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.4 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.5 | GO:1905031 | regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905031) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 1.0 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.9 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.4 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.6 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.6 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.4 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 1.8 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 2.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.9 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 1.2 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 1.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.9 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 1.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0009967 | positive regulation of signal transduction(GO:0009967) |
| 0.0 | 0.3 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.0 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 1.9 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.6 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.7 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.1 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.3 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.8 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.8 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |