Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
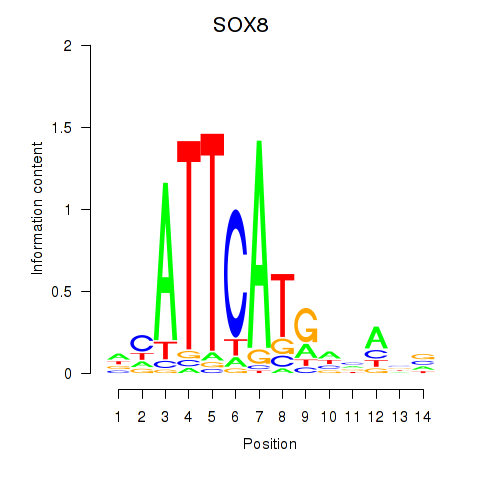
Results for SOX8
Z-value: 1.01
Transcription factors associated with SOX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX8
|
ENSG00000005513.9 | SOX8 |
Activity profile of SOX8 motif
Sorted Z-values of SOX8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_111415757 | 2.11 |
ENST00000429072.2 ENST00000271324.5 |
CD53 |
CD53 molecule |
| chr12_-_15104040 | 1.80 |
ENST00000541644.1 ENST00000545895.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr14_-_106471723 | 1.70 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr16_+_57392684 | 1.48 |
ENST00000219235.4 |
CCL22 |
chemokine (C-C motif) ligand 22 |
| chr5_+_89770696 | 1.30 |
ENST00000504930.1 ENST00000514483.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr5_+_89770664 | 1.21 |
ENST00000503973.1 ENST00000399107.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr18_-_11148587 | 1.11 |
ENST00000302079.6 ENST00000580640.1 ENST00000503781.3 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr6_-_32498046 | 1.07 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr2_+_102615416 | 1.05 |
ENST00000393414.2 |
IL1R2 |
interleukin 1 receptor, type II |
| chr16_+_29690358 | 1.04 |
ENST00000395384.4 ENST00000562473.1 |
QPRT |
quinolinate phosphoribosyltransferase |
| chr6_+_32605195 | 1.01 |
ENST00000374949.2 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr1_+_200993071 | 0.98 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr3_+_142342228 | 0.98 |
ENST00000337777.3 |
PLS1 |
plastin 1 |
| chr6_+_32605134 | 0.97 |
ENST00000343139.5 ENST00000395363.1 ENST00000496318.1 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
| chr4_+_40192656 | 0.97 |
ENST00000505618.1 |
RHOH |
ras homolog family member H |
| chr12_-_15103621 | 0.97 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr1_+_152956549 | 0.95 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chrX_-_132887729 | 0.94 |
ENST00000406757.2 |
GPC3 |
glypican 3 |
| chr19_-_47287990 | 0.93 |
ENST00000593713.1 ENST00000598022.1 ENST00000434726.2 |
SLC1A5 |
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr12_+_113354341 | 0.93 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_13516066 | 0.91 |
ENST00000332192.6 |
PRAMEF21 |
PRAME family member 21 |
| chr19_-_42806842 | 0.87 |
ENST00000596265.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr13_-_47012325 | 0.86 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr19_-_42806919 | 0.84 |
ENST00000595530.1 ENST00000538771.1 ENST00000601865.1 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr1_-_200992827 | 0.83 |
ENST00000332129.2 ENST00000422435.2 |
KIF21B |
kinesin family member 21B |
| chr7_+_64126535 | 0.82 |
ENST00000344930.3 |
ZNF107 |
zinc finger protein 107 |
| chr14_-_106642049 | 0.80 |
ENST00000390605.2 |
IGHV1-18 |
immunoglobulin heavy variable 1-18 |
| chr11_+_128563652 | 0.79 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr15_+_43985084 | 0.76 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr19_-_42806723 | 0.75 |
ENST00000262890.3 |
PAFAH1B3 |
platelet-activating factor acetylhydrolase 1b, catalytic subunit 3 (29kDa) |
| chr10_-_36813162 | 0.74 |
ENST00000440465.1 |
NAMPTL |
nicotinamide phosphoribosyltransferase-like |
| chr10_-_101825151 | 0.74 |
ENST00000441382.1 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chr15_+_43885252 | 0.73 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr5_-_78809950 | 0.73 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr16_+_87636474 | 0.72 |
ENST00000284262.2 |
JPH3 |
junctophilin 3 |
| chr1_+_70876926 | 0.69 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr12_-_10282742 | 0.68 |
ENST00000298523.5 ENST00000396484.2 ENST00000310002.4 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr18_-_29264669 | 0.66 |
ENST00000306851.5 |
B4GALT6 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr14_+_95078714 | 0.65 |
ENST00000393078.3 ENST00000393080.4 ENST00000467132.1 |
SERPINA3 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 |
| chr8_-_91095099 | 0.64 |
ENST00000265431.3 |
CALB1 |
calbindin 1, 28kDa |
| chr10_+_7745232 | 0.63 |
ENST00000358415.4 |
ITIH2 |
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr9_+_42671887 | 0.63 |
ENST00000456520.1 ENST00000377391.3 |
CBWD7 |
COBW domain containing 7 |
| chr12_-_123201337 | 0.62 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr11_-_57177586 | 0.60 |
ENST00000529411.1 |
RP11-872D17.8 |
Uncharacterized protein |
| chr18_+_61442629 | 0.58 |
ENST00000398019.2 ENST00000540675.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr14_-_106733624 | 0.58 |
ENST00000390610.2 |
IGHV1-24 |
immunoglobulin heavy variable 1-24 |
| chr1_-_152297679 | 0.57 |
ENST00000368799.1 |
FLG |
filaggrin |
| chr3_-_141719195 | 0.56 |
ENST00000397991.4 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr9_-_116837249 | 0.55 |
ENST00000466610.2 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr3_-_58196939 | 0.55 |
ENST00000394549.2 ENST00000461914.3 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr3_-_58196688 | 0.55 |
ENST00000486455.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr1_+_13742808 | 0.54 |
ENST00000602960.1 |
PRAMEF20 |
PRAME family member 20 |
| chr18_-_29264467 | 0.54 |
ENST00000383131.3 ENST00000237019.7 |
B4GALT6 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr1_-_153029980 | 0.54 |
ENST00000392653.2 |
SPRR2A |
small proline-rich protein 2A |
| chr2_+_74425689 | 0.52 |
ENST00000394053.2 ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr16_+_22308717 | 0.52 |
ENST00000299853.5 ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E |
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr7_+_100303676 | 0.48 |
ENST00000303151.4 |
POP7 |
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr14_+_35591858 | 0.47 |
ENST00000603544.1 |
KIAA0391 |
KIAA0391 |
| chr1_+_52082751 | 0.46 |
ENST00000447887.1 ENST00000435686.2 ENST00000428468.1 ENST00000453295.1 |
OSBPL9 |
oxysterol binding protein-like 9 |
| chr1_+_153330322 | 0.45 |
ENST00000368738.3 |
S100A9 |
S100 calcium binding protein A9 |
| chr6_-_32160622 | 0.45 |
ENST00000487761.1 ENST00000375040.3 |
GPSM3 |
G-protein signaling modulator 3 |
| chr14_+_39583427 | 0.43 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr3_-_54962100 | 0.43 |
ENST00000273286.5 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr4_+_155484155 | 0.43 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr16_+_30075783 | 0.42 |
ENST00000412304.2 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr4_-_145061788 | 0.42 |
ENST00000512064.1 ENST00000512789.1 ENST00000504786.1 ENST00000503627.1 ENST00000535709.1 ENST00000324022.10 ENST00000360771.4 ENST00000283126.7 |
GYPA GYPB |
glycophorin A (MNS blood group) glycophorin B (MNS blood group) |
| chr14_-_106967788 | 0.41 |
ENST00000390622.2 |
IGHV1-46 |
immunoglobulin heavy variable 1-46 |
| chr17_-_34207295 | 0.41 |
ENST00000463941.1 ENST00000293272.3 |
CCL5 |
chemokine (C-C motif) ligand 5 |
| chr14_+_35591928 | 0.41 |
ENST00000605870.1 ENST00000557404.3 |
KIAA0391 |
KIAA0391 |
| chr16_+_30075463 | 0.40 |
ENST00000562168.1 ENST00000569545.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr14_-_107049312 | 0.40 |
ENST00000390627.2 |
IGHV3-53 |
immunoglobulin heavy variable 3-53 |
| chr12_-_123187890 | 0.40 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr1_+_161475208 | 0.40 |
ENST00000367972.4 ENST00000271450.6 |
FCGR2A |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
| chr14_+_88471468 | 0.39 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr11_+_17281900 | 0.39 |
ENST00000530527.1 |
NUCB2 |
nucleobindin 2 |
| chr6_-_22297730 | 0.38 |
ENST00000306482.1 |
PRL |
prolactin |
| chr1_+_26496362 | 0.38 |
ENST00000374266.5 ENST00000270812.5 |
ZNF593 |
zinc finger protein 593 |
| chr12_-_10282836 | 0.38 |
ENST00000304084.8 ENST00000353231.5 ENST00000525605.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr4_-_100356291 | 0.38 |
ENST00000476959.1 ENST00000482593.1 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chrX_+_15767971 | 0.37 |
ENST00000479740.1 ENST00000454127.2 |
CA5B |
carbonic anhydrase VB, mitochondrial |
| chr18_-_3845321 | 0.37 |
ENST00000539435.1 ENST00000400147.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr15_-_58306295 | 0.37 |
ENST00000559517.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr19_-_45681482 | 0.37 |
ENST00000592647.1 ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A |
trafficking protein particle complex 6A |
| chr4_-_144940477 | 0.37 |
ENST00000513128.1 ENST00000429670.2 ENST00000502664.1 |
GYPB |
glycophorin B (MNS blood group) |
| chr12_-_11548496 | 0.36 |
ENST00000389362.4 ENST00000565533.1 ENST00000546254.1 |
PRB2 PRB1 |
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr11_+_128563948 | 0.36 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_155484103 | 0.36 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr17_+_41132564 | 0.36 |
ENST00000361677.1 ENST00000589705.1 |
RUNDC1 |
RUN domain containing 1 |
| chr9_+_2157655 | 0.36 |
ENST00000452193.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_11674213 | 0.35 |
ENST00000381486.2 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr3_+_52828805 | 0.35 |
ENST00000416872.2 ENST00000449956.2 |
ITIH3 |
inter-alpha-trypsin inhibitor heavy chain 3 |
| chr17_-_41132410 | 0.35 |
ENST00000409446.3 ENST00000453594.1 ENST00000409399.1 ENST00000421990.2 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr14_+_35591735 | 0.35 |
ENST00000604948.1 ENST00000605201.1 ENST00000250377.7 ENST00000321130.10 ENST00000534898.4 |
KIAA0391 |
KIAA0391 |
| chr20_+_32150140 | 0.34 |
ENST00000344201.3 ENST00000346541.3 ENST00000397800.1 ENST00000397798.2 ENST00000492345.1 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
| chr17_-_47723943 | 0.34 |
ENST00000510476.1 ENST00000503676.1 |
SPOP |
speckle-type POZ protein |
| chr2_+_204801471 | 0.34 |
ENST00000316386.6 ENST00000435193.1 |
ICOS |
inducible T-cell co-stimulator |
| chr12_+_28410128 | 0.33 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr16_-_30122717 | 0.33 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr2_+_97203082 | 0.33 |
ENST00000454558.2 |
ARID5A |
AT rich interactive domain 5A (MRF1-like) |
| chr8_-_20040638 | 0.32 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr8_+_21915368 | 0.32 |
ENST00000265800.5 ENST00000517418.1 |
DMTN |
dematin actin binding protein |
| chr15_-_38519066 | 0.32 |
ENST00000561320.1 ENST00000561161.1 |
RP11-346D14.1 |
RP11-346D14.1 |
| chr5_+_54455946 | 0.32 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr2_+_166326157 | 0.32 |
ENST00000421875.1 ENST00000314499.7 ENST00000409664.1 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
| chr5_-_160973649 | 0.31 |
ENST00000393959.1 ENST00000517547.1 |
GABRB2 |
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr11_-_128457446 | 0.31 |
ENST00000392668.4 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
| chr4_-_144826682 | 0.30 |
ENST00000358615.4 ENST00000437468.2 |
GYPE |
glycophorin E (MNS blood group) |
| chr7_-_32529973 | 0.30 |
ENST00000410044.1 ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5 |
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr15_-_20170354 | 0.29 |
ENST00000338912.5 |
IGHV1OR15-9 |
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr3_-_52090461 | 0.29 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr8_-_86253888 | 0.29 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr8_+_107738240 | 0.29 |
ENST00000449762.2 ENST00000297447.6 |
OXR1 |
oxidation resistance 1 |
| chr7_+_139478030 | 0.29 |
ENST00000425687.1 ENST00000263552.6 ENST00000438104.1 ENST00000336425.5 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
| chr1_+_235492300 | 0.29 |
ENST00000476121.1 ENST00000497327.1 |
GGPS1 |
geranylgeranyl diphosphate synthase 1 |
| chr16_+_30075595 | 0.28 |
ENST00000563060.2 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chr18_-_3845293 | 0.28 |
ENST00000400145.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr3_-_49459878 | 0.28 |
ENST00000546031.1 ENST00000458307.2 ENST00000430521.1 |
AMT |
aminomethyltransferase |
| chr6_-_136788001 | 0.28 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr2_-_89521942 | 0.28 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr13_+_97928395 | 0.27 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr16_+_31271274 | 0.26 |
ENST00000287497.8 ENST00000544665.3 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
| chr12_+_21207503 | 0.26 |
ENST00000545916.1 |
SLCO1B7 |
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr11_-_46142615 | 0.25 |
ENST00000529734.1 ENST00000323180.6 |
PHF21A |
PHD finger protein 21A |
| chr17_+_1646130 | 0.25 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr3_-_49851313 | 0.25 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr4_+_89299994 | 0.24 |
ENST00000264346.7 |
HERC6 |
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr1_+_24019099 | 0.24 |
ENST00000443624.1 ENST00000458455.1 |
RPL11 |
ribosomal protein L11 |
| chr1_+_52682052 | 0.24 |
ENST00000371591.1 |
ZFYVE9 |
zinc finger, FYVE domain containing 9 |
| chr8_+_38758737 | 0.24 |
ENST00000521746.1 ENST00000420274.1 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
| chr18_+_56892724 | 0.23 |
ENST00000456142.3 ENST00000530323.1 |
GRP |
gastrin-releasing peptide |
| chr3_-_176914238 | 0.23 |
ENST00000430069.1 ENST00000428970.1 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
| chr12_+_56325812 | 0.23 |
ENST00000394147.1 ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA |
diacylglycerol kinase, alpha 80kDa |
| chr11_+_59522837 | 0.23 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chrX_-_53461288 | 0.23 |
ENST00000375298.4 ENST00000375304.5 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr1_-_222763240 | 0.23 |
ENST00000352967.4 ENST00000391882.1 ENST00000543857.1 |
TAF1A |
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa |
| chrX_+_100474906 | 0.22 |
ENST00000541709.1 |
DRP2 |
dystrophin related protein 2 |
| chr4_-_100356551 | 0.22 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr19_-_6333614 | 0.21 |
ENST00000301452.4 |
ACER1 |
alkaline ceramidase 1 |
| chr6_-_27100529 | 0.21 |
ENST00000607124.1 ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ |
histone cluster 1, H2bj |
| chr22_-_36903069 | 0.21 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr3_+_49711391 | 0.21 |
ENST00000296456.5 ENST00000449966.1 |
APEH |
acylaminoacyl-peptide hydrolase |
| chr3_-_21792838 | 0.21 |
ENST00000281523.2 |
ZNF385D |
zinc finger protein 385D |
| chr20_-_44176013 | 0.21 |
ENST00000555685.1 |
EPPIN |
epididymal peptidase inhibitor |
| chr11_+_60995849 | 0.21 |
ENST00000537932.1 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr9_-_128246769 | 0.21 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr6_+_63921351 | 0.21 |
ENST00000370659.1 |
FKBP1C |
FK506 binding protein 1C |
| chr3_-_49459865 | 0.20 |
ENST00000427987.1 |
AMT |
aminomethyltransferase |
| chrX_+_49091920 | 0.20 |
ENST00000376227.3 |
CCDC22 |
coiled-coil domain containing 22 |
| chr6_-_133079022 | 0.20 |
ENST00000525289.1 ENST00000326499.6 |
VNN2 |
vanin 2 |
| chr2_-_163008903 | 0.20 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr7_-_36634181 | 0.20 |
ENST00000538464.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr16_-_28503357 | 0.19 |
ENST00000333496.9 ENST00000561505.1 ENST00000567963.1 ENST00000354630.5 ENST00000355477.5 ENST00000357076.5 ENST00000565688.1 ENST00000359984.7 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
| chr1_-_67142710 | 0.19 |
ENST00000502413.2 |
AL139147.1 |
Uncharacterized protein |
| chr17_+_2240916 | 0.19 |
ENST00000574563.1 |
SGSM2 |
small G protein signaling modulator 2 |
| chr5_-_160279207 | 0.18 |
ENST00000327245.5 |
ATP10B |
ATPase, class V, type 10B |
| chrX_-_53461305 | 0.18 |
ENST00000168216.6 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr4_-_38858428 | 0.17 |
ENST00000436693.2 ENST00000508254.1 ENST00000514655.1 ENST00000506146.1 |
TLR6 TLR1 |
toll-like receptor 6 toll-like receptor 1 |
| chr14_-_107283278 | 0.17 |
ENST00000390639.2 |
IGHV7-81 |
immunoglobulin heavy variable 7-81 (non-functional) |
| chr6_-_7313381 | 0.17 |
ENST00000489567.1 ENST00000479365.1 ENST00000462112.1 ENST00000397511.2 ENST00000534851.1 ENST00000474597.1 ENST00000244763.4 |
SSR1 |
signal sequence receptor, alpha |
| chr18_+_32173276 | 0.17 |
ENST00000591816.1 ENST00000588125.1 ENST00000598334.1 ENST00000588684.1 ENST00000554864.3 ENST00000399121.5 ENST00000595022.1 ENST00000269190.7 ENST00000399097.3 |
DTNA |
dystrobrevin, alpha |
| chr16_+_82068830 | 0.17 |
ENST00000199936.4 |
HSD17B2 |
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr5_-_94417339 | 0.16 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr8_+_7752151 | 0.16 |
ENST00000302247.2 |
DEFB4A |
defensin, beta 4A |
| chr11_+_10477733 | 0.16 |
ENST00000528723.1 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr6_+_118869452 | 0.16 |
ENST00000357525.5 |
PLN |
phospholamban |
| chr1_+_192605252 | 0.16 |
ENST00000391995.2 ENST00000543215.1 |
RGS13 |
regulator of G-protein signaling 13 |
| chr9_+_127624387 | 0.16 |
ENST00000353214.2 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
| chr1_-_13117736 | 0.16 |
ENST00000376192.5 ENST00000376182.1 |
PRAMEF6 |
PRAME family member 6 |
| chrX_+_18725758 | 0.16 |
ENST00000472826.1 ENST00000544635.1 ENST00000496075.2 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
| chr5_+_175288631 | 0.16 |
ENST00000509837.1 |
CPLX2 |
complexin 2 |
| chr7_+_23145366 | 0.16 |
ENST00000339077.5 ENST00000322275.5 ENST00000539124.1 ENST00000542558.1 |
KLHL7 |
kelch-like family member 7 |
| chr19_+_51728316 | 0.15 |
ENST00000436584.2 ENST00000421133.2 ENST00000391796.3 ENST00000262262.4 |
CD33 |
CD33 molecule |
| chr10_-_1779663 | 0.15 |
ENST00000381312.1 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr4_+_95972822 | 0.15 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr3_+_185080908 | 0.15 |
ENST00000265026.3 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chr3_+_173116225 | 0.15 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr16_+_87985029 | 0.15 |
ENST00000439677.1 ENST00000286122.7 ENST00000355163.5 ENST00000454563.1 ENST00000479780.2 ENST00000393208.2 ENST00000412691.1 ENST00000355022.4 |
BANP |
BTG3 associated nuclear protein |
| chr17_-_8021710 | 0.15 |
ENST00000380149.1 ENST00000448843.2 |
ALOXE3 |
arachidonate lipoxygenase 3 |
| chr2_+_204732666 | 0.14 |
ENST00000295854.6 ENST00000472206.1 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
| chr3_+_38347427 | 0.13 |
ENST00000273173.4 |
SLC22A14 |
solute carrier family 22, member 14 |
| chr7_-_14942283 | 0.13 |
ENST00000402815.1 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr1_-_116383738 | 0.13 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr2_+_204732487 | 0.13 |
ENST00000302823.3 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
| chr15_-_89755034 | 0.13 |
ENST00000563254.1 |
RLBP1 |
retinaldehyde binding protein 1 |
| chr1_+_52870227 | 0.13 |
ENST00000257181.9 |
PRPF38A |
pre-mRNA processing factor 38A |
| chr22_-_43411106 | 0.13 |
ENST00000453643.1 ENST00000263246.3 ENST00000337959.4 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
| chr6_-_159421198 | 0.12 |
ENST00000252655.1 ENST00000297262.3 ENST00000367069.2 |
RSPH3 |
radial spoke 3 homolog (Chlamydomonas) |
| chr15_-_66649010 | 0.12 |
ENST00000367709.4 ENST00000261881.4 |
TIPIN |
TIMELESS interacting protein |
| chr8_+_26150628 | 0.12 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr20_-_44176060 | 0.12 |
ENST00000354280.4 ENST00000504988.1 |
EPPIN EPPIN-WFDC6 |
epididymal peptidase inhibitor EPPIN-WFDC6 readthrough |
| chr16_+_57769635 | 0.12 |
ENST00000379661.3 ENST00000562592.1 ENST00000566726.1 |
KATNB1 |
katanin p80 (WD repeat containing) subunit B 1 |
| chr1_-_13115578 | 0.12 |
ENST00000414205.2 |
PRAMEF6 |
PRAME family member 6 |
| chr11_-_8680383 | 0.12 |
ENST00000299550.6 |
TRIM66 |
tripartite motif containing 66 |
| chr18_-_78005231 | 0.12 |
ENST00000470488.2 ENST00000353265.3 |
PARD6G |
par-6 family cell polarity regulator gamma |
| chr17_-_19648683 | 0.12 |
ENST00000573368.1 ENST00000457500.2 |
ALDH3A1 |
aldehyde dehydrogenase 3 family, member A1 |
| chr3_+_148545586 | 0.12 |
ENST00000282957.4 ENST00000468341.1 |
CPB1 |
carboxypeptidase B1 (tissue) |
| chr22_-_42466782 | 0.11 |
ENST00000396398.3 ENST00000403363.1 ENST00000402937.1 |
NAGA |
N-acetylgalactosaminidase, alpha- |
| chr22_-_29784519 | 0.11 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr10_-_6104253 | 0.11 |
ENST00000256876.6 ENST00000379954.1 ENST00000379959.3 |
IL2RA |
interleukin 2 receptor, alpha |
| chr2_+_113816215 | 0.10 |
ENST00000346807.3 |
IL36RN |
interleukin 36 receptor antagonist |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.3 | 1.6 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.3 | 1.1 | GO:0050712 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of interleukin-1 alpha secretion(GO:0050712) |
| 0.2 | 0.7 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.2 | 0.9 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.2 | 0.9 | GO:1903803 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.2 | 1.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.2 | 0.6 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.2 | 1.0 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.7 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.2 | 0.5 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.7 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.6 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.3 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.1 | 2.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 2.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.4 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.8 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.6 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.1 | 0.2 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.1 | 1.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.2 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.2 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.1 | 4.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.1 | 0.3 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 | 0.3 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.1 | 1.1 | GO:0010623 | programmed cell death involved in cell development(GO:0010623) |
| 0.1 | 0.3 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.2 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.1 | 0.3 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.1 | 1.1 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 0.4 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 1.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:1901318 | negative regulation of sperm motility(GO:1901318) |
| 0.0 | 0.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.0 | 1.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.5 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.3 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.4 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.6 | GO:0018298 | porphyrin-containing compound catabolic process(GO:0006787) protein-chromophore linkage(GO:0018298) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 2.1 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.0 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 3.4 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0001777 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.7 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 1.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.1 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 1.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.3 | GO:0046514 | ceramide catabolic process(GO:0046514) |
| 0.0 | 1.0 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 0.1 | GO:0015888 | thiamine transport(GO:0015888) thiamine transmembrane transport(GO:0071934) |
| 0.0 | 1.0 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.1 | GO:0044854 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.4 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.0 | 0.3 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.4 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.3 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.2 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.6 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.9 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0072584 | caveolin-mediated endocytosis(GO:0072584) |
| 0.0 | 1.1 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.5 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0071314 | cellular response to cocaine(GO:0071314) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.2 | 0.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 1.0 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 3.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 3.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 3.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.2 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.4 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.3 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 3.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.3 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.2 | GO:0097486 | multivesicular body lumen(GO:0097486) |
| 0.0 | 0.4 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 2.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.1 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.6 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.9 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.5 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.3 | 1.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.3 | 2.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 0.9 | GO:0060422 | peptidyl-dipeptidase inhibitor activity(GO:0060422) |
| 0.2 | 1.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.2 | 1.1 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 0.5 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.6 | GO:0035276 | aldehyde oxidase activity(GO:0004031) ethanol binding(GO:0035276) |
| 0.1 | 2.0 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 0.6 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.5 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.1 | 1.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 1.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 1.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.1 | 1.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.2 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.1 | 3.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.4 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.3 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 2.9 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.1 | 0.2 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.7 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 1.0 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 1.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.0 | 1.1 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.7 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0050542 | icosanoid binding(GO:0050542) arachidonic acid binding(GO:0050544) fatty acid derivative binding(GO:1901567) |
| 0.0 | 2.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.3 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 1.1 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0015403 | thiamine transmembrane transporter activity(GO:0015234) thiamine uptake transmembrane transporter activity(GO:0015403) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.3 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.3 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 0.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 1.3 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |