Project
ENCODE cell lines, expression (Ernst 2011)
Navigation
Downloads
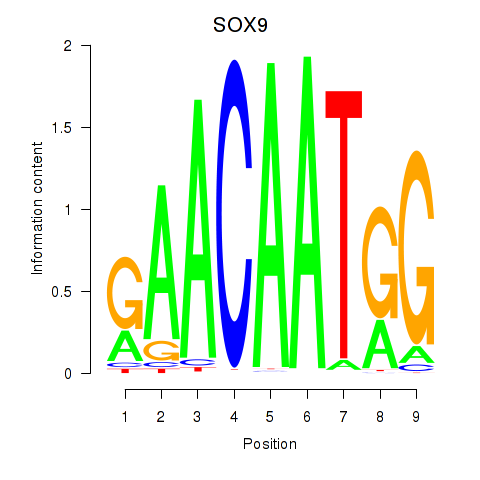
Results for SOX9
Z-value: 1.16
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SOX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| SOX9 | hg19_v2_chr17_+_70117153_70117174 | -0.03 | 9.1e-01 | Click! |
Activity profile of SOX9 motif
Sorted Z-values of SOX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of SOX9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_128841503 | 3.50 |
ENST00000368215.3 ENST00000532331.1 ENST00000368213.5 ENST00000368207.3 ENST00000525459.1 ENST00000368210.3 ENST00000368226.4 ENST00000368227.3 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
| chr12_-_6233828 | 3.25 |
ENST00000572068.1 ENST00000261405.5 |
VWF |
von Willebrand factor |
| chrX_-_38080077 | 2.92 |
ENST00000378533.3 ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX |
sushi-repeat containing protein, X-linked |
| chr5_-_16936340 | 2.92 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr7_+_134464414 | 2.70 |
ENST00000361901.2 |
CALD1 |
caldesmon 1 |
| chr7_-_27205136 | 2.27 |
ENST00000396345.1 ENST00000343483.6 |
HOXA9 |
homeobox A9 |
| chr2_-_151344172 | 2.25 |
ENST00000375734.2 ENST00000263895.4 ENST00000454202.1 |
RND3 |
Rho family GTPase 3 |
| chr7_+_134464376 | 2.12 |
ENST00000454108.1 ENST00000361675.2 |
CALD1 |
caldesmon 1 |
| chr1_-_68698222 | 2.08 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr2_-_1748214 | 2.00 |
ENST00000433670.1 ENST00000425171.1 ENST00000252804.4 |
PXDN |
peroxidasin homolog (Drosophila) |
| chr7_-_131241361 | 1.76 |
ENST00000378555.3 ENST00000322985.9 ENST00000541194.1 ENST00000537928.1 |
PODXL |
podocalyxin-like |
| chr5_-_146833485 | 1.66 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr3_-_149688896 | 1.58 |
ENST00000239940.7 |
PFN2 |
profilin 2 |
| chr3_-_149688502 | 1.58 |
ENST00000481767.1 ENST00000475518.1 |
PFN2 |
profilin 2 |
| chr7_-_11871815 | 1.56 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr9_+_116263778 | 1.53 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr9_+_116263639 | 1.52 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr17_-_39743139 | 1.44 |
ENST00000167586.6 |
KRT14 |
keratin 14 |
| chr3_-_149688655 | 1.39 |
ENST00000461930.1 ENST00000423691.2 ENST00000490975.1 ENST00000461868.1 ENST00000452853.2 |
PFN2 |
profilin 2 |
| chr1_-_79472365 | 1.37 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr7_+_16793160 | 1.36 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr5_+_76011868 | 1.36 |
ENST00000319211.4 |
F2R |
coagulation factor II (thrombin) receptor |
| chr5_-_39425222 | 1.35 |
ENST00000320816.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_-_107643674 | 1.34 |
ENST00000222399.6 |
LAMB1 |
laminin, beta 1 |
| chr4_-_186732048 | 1.33 |
ENST00000448662.2 ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr14_+_85996471 | 1.31 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr1_+_93913713 | 1.30 |
ENST00000604705.1 ENST00000370253.2 |
FNBP1L |
formin binding protein 1-like |
| chr16_+_66400533 | 1.30 |
ENST00000341529.3 |
CDH5 |
cadherin 5, type 2 (vascular endothelium) |
| chr5_-_39425290 | 1.29 |
ENST00000545653.1 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr16_+_3068393 | 1.26 |
ENST00000573001.1 |
TNFRSF12A |
tumor necrosis factor receptor superfamily, member 12A |
| chr6_+_148663729 | 1.23 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr6_+_114178512 | 1.23 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr7_-_111846435 | 1.21 |
ENST00000437633.1 ENST00000428084.1 |
DOCK4 |
dedicator of cytokinesis 4 |
| chr1_-_68698197 | 1.19 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr12_-_52887034 | 1.17 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr22_-_19512893 | 1.11 |
ENST00000403084.1 ENST00000413119.2 |
CLDN5 |
claudin 5 |
| chr12_-_10251603 | 1.08 |
ENST00000457018.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr18_+_3449821 | 1.04 |
ENST00000407501.2 ENST00000405385.3 ENST00000546979.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr12_-_10251576 | 1.02 |
ENST00000315330.4 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr2_+_102314161 | 1.02 |
ENST00000425019.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr12_-_10251539 | 1.01 |
ENST00000420265.2 |
CLEC1A |
C-type lectin domain family 1, member A |
| chr1_+_43766642 | 1.01 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr2_+_217498105 | 1.01 |
ENST00000233809.4 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr22_+_38142235 | 1.01 |
ENST00000407319.2 ENST00000403663.2 ENST00000428075.1 |
TRIOBP |
TRIO and F-actin binding protein |
| chr10_-_105845674 | 0.99 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr12_+_49212514 | 0.97 |
ENST00000301050.2 ENST00000548279.1 ENST00000547230.1 |
CACNB3 |
calcium channel, voltage-dependent, beta 3 subunit |
| chr5_-_39425068 | 0.96 |
ENST00000515700.1 ENST00000339788.6 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr8_-_82395461 | 0.96 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr2_-_175869936 | 0.95 |
ENST00000409900.3 |
CHN1 |
chimerin 1 |
| chr14_+_85996507 | 0.92 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr4_-_2264015 | 0.91 |
ENST00000337190.2 |
MXD4 |
MAX dimerization protein 4 |
| chr4_+_175204818 | 0.90 |
ENST00000503780.1 |
CEP44 |
centrosomal protein 44kDa |
| chr8_+_79578282 | 0.89 |
ENST00000263849.4 |
ZC2HC1A |
zinc finger, C2HC-type containing 1A |
| chr18_-_21891460 | 0.89 |
ENST00000357041.4 |
OSBPL1A |
oxysterol binding protein-like 1A |
| chr1_-_225840747 | 0.88 |
ENST00000366843.2 ENST00000366844.3 |
ENAH |
enabled homolog (Drosophila) |
| chr15_+_71184931 | 0.87 |
ENST00000560369.1 ENST00000260382.5 |
LRRC49 |
leucine rich repeat containing 49 |
| chr12_-_71003568 | 0.87 |
ENST00000547715.1 ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
| chr5_+_15500280 | 0.84 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr5_-_146889619 | 0.83 |
ENST00000343218.5 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr2_-_55277692 | 0.80 |
ENST00000394611.2 |
RTN4 |
reticulon 4 |
| chr18_+_3451646 | 0.78 |
ENST00000345133.5 ENST00000330513.5 ENST00000549546.1 |
TGIF1 |
TGFB-induced factor homeobox 1 |
| chr8_+_26435359 | 0.77 |
ENST00000311151.5 |
DPYSL2 |
dihydropyrimidinase-like 2 |
| chr18_+_7946941 | 0.77 |
ENST00000444013.1 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chr2_-_55277654 | 0.75 |
ENST00000337526.6 ENST00000317610.7 ENST00000357732.4 |
RTN4 |
reticulon 4 |
| chr11_+_77532233 | 0.75 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr3_+_113616317 | 0.74 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chr8_-_93115445 | 0.72 |
ENST00000523629.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr18_+_56530794 | 0.71 |
ENST00000590285.1 ENST00000586085.1 ENST00000589288.1 |
ZNF532 |
zinc finger protein 532 |
| chr12_-_95611149 | 0.70 |
ENST00000549499.1 ENST00000343958.4 ENST00000546711.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr5_+_40679584 | 0.70 |
ENST00000302472.3 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
| chr7_+_77167376 | 0.70 |
ENST00000435495.2 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
| chr7_+_18535346 | 0.70 |
ENST00000405010.3 ENST00000406451.4 ENST00000428307.2 |
HDAC9 |
histone deacetylase 9 |
| chr2_-_55277512 | 0.70 |
ENST00000402434.2 |
RTN4 |
reticulon 4 |
| chr2_-_55277436 | 0.69 |
ENST00000354474.6 |
RTN4 |
reticulon 4 |
| chr11_+_60691924 | 0.69 |
ENST00000544065.1 ENST00000453848.2 ENST00000005286.4 |
TMEM132A |
transmembrane protein 132A |
| chr18_-_53177984 | 0.69 |
ENST00000543082.1 |
TCF4 |
transcription factor 4 |
| chr17_-_7297519 | 0.68 |
ENST00000576362.1 ENST00000571078.1 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr19_+_16187816 | 0.66 |
ENST00000588410.1 |
TPM4 |
tropomyosin 4 |
| chr9_+_103235365 | 0.66 |
ENST00000374879.4 |
TMEFF1 |
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr1_+_66458072 | 0.66 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr3_-_57583052 | 0.65 |
ENST00000496292.1 ENST00000489843.1 |
ARF4 |
ADP-ribosylation factor 4 |
| chr3_+_43328004 | 0.65 |
ENST00000454177.1 ENST00000429705.2 ENST00000296088.7 ENST00000437827.1 |
SNRK |
SNF related kinase |
| chr12_-_8815299 | 0.63 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr1_+_155293702 | 0.62 |
ENST00000368347.4 |
RUSC1 |
RUN and SH3 domain containing 1 |
| chr22_-_22292934 | 0.62 |
ENST00000538191.1 ENST00000424647.1 ENST00000407142.1 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr1_+_155294342 | 0.62 |
ENST00000292254.4 |
RUSC1 |
RUN and SH3 domain containing 1 |
| chr9_+_27109133 | 0.62 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr3_-_57583130 | 0.61 |
ENST00000303436.6 |
ARF4 |
ADP-ribosylation factor 4 |
| chr19_+_48972459 | 0.61 |
ENST00000427476.1 |
CYTH2 |
cytohesin 2 |
| chr15_+_85923797 | 0.61 |
ENST00000559362.1 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
| chr7_+_12727250 | 0.61 |
ENST00000404894.1 |
ARL4A |
ADP-ribosylation factor-like 4A |
| chr17_-_7297833 | 0.59 |
ENST00000571802.1 ENST00000576201.1 ENST00000573213.1 ENST00000324822.11 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr19_-_44124019 | 0.59 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr3_+_51422478 | 0.59 |
ENST00000528157.1 |
MANF |
mesencephalic astrocyte-derived neurotrophic factor |
| chr5_-_146833222 | 0.58 |
ENST00000534907.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr20_-_17662878 | 0.57 |
ENST00000377813.1 ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1 |
ribosome binding protein 1 |
| chr12_-_8815215 | 0.57 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr1_+_155294264 | 0.57 |
ENST00000368349.4 |
RUSC1 |
RUN and SH3 domain containing 1 |
| chr7_+_116312411 | 0.56 |
ENST00000456159.1 ENST00000397752.3 ENST00000318493.6 |
MET |
met proto-oncogene |
| chr5_+_140739537 | 0.55 |
ENST00000522605.1 |
PCDHGB2 |
protocadherin gamma subfamily B, 2 |
| chr4_+_78078304 | 0.55 |
ENST00000316355.5 ENST00000354403.5 ENST00000502280.1 |
CCNG2 |
cyclin G2 |
| chr11_+_77532155 | 0.54 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr10_+_71812552 | 0.54 |
ENST00000373255.4 |
H2AFY2 |
H2A histone family, member Y2 |
| chr17_+_8213590 | 0.53 |
ENST00000361926.3 |
ARHGEF15 |
Rho guanine nucleotide exchange factor (GEF) 15 |
| chr4_+_154125565 | 0.52 |
ENST00000338700.5 |
TRIM2 |
tripartite motif containing 2 |
| chr2_-_55276320 | 0.52 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr7_-_27219849 | 0.49 |
ENST00000396344.4 |
HOXA10 |
homeobox A10 |
| chr14_+_59655369 | 0.48 |
ENST00000360909.3 ENST00000351081.1 ENST00000556135.1 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
| chr3_-_123339418 | 0.48 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr16_+_15528332 | 0.47 |
ENST00000566490.1 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr5_+_140729649 | 0.47 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr11_-_6677018 | 0.47 |
ENST00000299441.3 |
DCHS1 |
dachsous cadherin-related 1 |
| chr5_+_140593509 | 0.47 |
ENST00000341948.4 |
PCDHB13 |
protocadherin beta 13 |
| chrY_+_15016013 | 0.46 |
ENST00000360160.4 ENST00000454054.1 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
| chr1_+_200708671 | 0.45 |
ENST00000358823.2 |
CAMSAP2 |
calmodulin regulated spectrin-associated protein family, member 2 |
| chr16_-_89556942 | 0.45 |
ENST00000301030.4 |
ANKRD11 |
ankyrin repeat domain 11 |
| chr13_+_98795434 | 0.45 |
ENST00000376586.2 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr13_+_98795505 | 0.45 |
ENST00000319562.6 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr10_+_102106829 | 0.45 |
ENST00000370355.2 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr6_-_11232891 | 0.45 |
ENST00000379433.5 ENST00000379446.5 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr3_-_123339343 | 0.45 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr4_-_149363662 | 0.44 |
ENST00000355292.3 ENST00000358102.3 |
NR3C2 |
nuclear receptor subfamily 3, group C, member 2 |
| chr17_-_73505961 | 0.43 |
ENST00000433559.2 |
CASKIN2 |
CASK interacting protein 2 |
| chr8_+_102504651 | 0.43 |
ENST00000251808.3 ENST00000521085.1 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
| chr20_-_17662705 | 0.43 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chr5_-_141338627 | 0.42 |
ENST00000231484.3 |
PCDH12 |
protocadherin 12 |
| chr12_-_49582978 | 0.41 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr5_+_74633036 | 0.41 |
ENST00000343975.5 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr11_+_128563948 | 0.41 |
ENST00000534087.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr6_+_56954867 | 0.41 |
ENST00000370708.4 ENST00000370702.1 |
ZNF451 |
zinc finger protein 451 |
| chr1_+_25071848 | 0.40 |
ENST00000374379.4 |
CLIC4 |
chloride intracellular channel 4 |
| chr3_-_88108212 | 0.39 |
ENST00000482016.1 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr7_+_26191809 | 0.38 |
ENST00000056233.3 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
| chr5_+_140797296 | 0.38 |
ENST00000398594.2 |
PCDHGB7 |
protocadherin gamma subfamily B, 7 |
| chr20_-_43150601 | 0.37 |
ENST00000541235.1 ENST00000255175.1 ENST00000342374.4 |
SERINC3 |
serine incorporator 3 |
| chr1_+_64239657 | 0.37 |
ENST00000371080.1 ENST00000371079.1 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
| chr18_-_34408902 | 0.37 |
ENST00000593035.1 ENST00000383056.3 ENST00000588909.1 ENST00000590337.1 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr7_+_16685756 | 0.37 |
ENST00000415365.1 ENST00000258761.3 ENST00000433922.2 ENST00000452975.2 ENST00000405202.1 |
BZW2 |
basic leucine zipper and W2 domains 2 |
| chrX_-_45060135 | 0.36 |
ENST00000398000.2 ENST00000377934.4 |
CXorf36 |
chromosome X open reading frame 36 |
| chr5_+_74632993 | 0.36 |
ENST00000287936.4 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
| chr12_+_53443680 | 0.36 |
ENST00000314250.6 ENST00000451358.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr19_-_8408139 | 0.36 |
ENST00000330915.3 ENST00000593649.1 ENST00000595639.1 |
KANK3 |
KN motif and ankyrin repeat domains 3 |
| chr1_+_164528866 | 0.35 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr11_+_13299186 | 0.35 |
ENST00000527998.1 ENST00000396441.3 ENST00000533520.1 ENST00000529825.1 ENST00000389707.4 ENST00000401424.1 ENST00000529388.1 ENST00000530357.1 ENST00000403290.1 ENST00000361003.4 ENST00000389708.3 ENST00000403510.3 ENST00000482049.1 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
| chr11_+_131781290 | 0.35 |
ENST00000425719.2 ENST00000374784.1 |
NTM |
neurotrimin |
| chr14_-_55878538 | 0.35 |
ENST00000247178.5 |
ATG14 |
autophagy related 14 |
| chr7_+_128379346 | 0.34 |
ENST00000535011.2 ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU |
calumenin |
| chr14_-_88459503 | 0.34 |
ENST00000393568.4 ENST00000261304.2 |
GALC |
galactosylceramidase |
| chr8_+_70378852 | 0.34 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr9_+_27109392 | 0.34 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr11_-_75236867 | 0.34 |
ENST00000376282.3 ENST00000336898.3 |
GDPD5 |
glycerophosphodiester phosphodiesterase domain containing 5 |
| chr10_+_70748487 | 0.33 |
ENST00000361983.4 |
KIAA1279 |
KIAA1279 |
| chr5_+_140767452 | 0.33 |
ENST00000519479.1 |
PCDHGB4 |
protocadherin gamma subfamily B, 4 |
| chr22_+_38864041 | 0.32 |
ENST00000216014.4 ENST00000409006.3 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr3_+_49507559 | 0.32 |
ENST00000421560.1 ENST00000308775.2 ENST00000545947.1 ENST00000541308.1 ENST00000539901.1 ENST00000538711.1 ENST00000418588.1 |
DAG1 |
dystroglycan 1 (dystrophin-associated glycoprotein 1) |
| chr16_+_14802801 | 0.32 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr18_-_34409116 | 0.31 |
ENST00000334295.4 |
TPGS2 |
tubulin polyglutamylase complex subunit 2 |
| chr3_+_25469802 | 0.31 |
ENST00000330688.4 |
RARB |
retinoic acid receptor, beta |
| chr20_+_42086525 | 0.29 |
ENST00000244020.3 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
| chr19_+_42580274 | 0.29 |
ENST00000359044.4 |
ZNF574 |
zinc finger protein 574 |
| chr1_-_21503337 | 0.29 |
ENST00000400422.1 ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr16_-_15474904 | 0.29 |
ENST00000534094.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr3_-_52001448 | 0.29 |
ENST00000461554.1 ENST00000395013.3 ENST00000428823.2 ENST00000483411.1 ENST00000461544.1 ENST00000355852.2 |
PCBP4 |
poly(rC) binding protein 4 |
| chr15_-_59041768 | 0.29 |
ENST00000402627.1 ENST00000396140.2 ENST00000559053.1 ENST00000561288.1 |
ADAM10 |
ADAM metallopeptidase domain 10 |
| chr19_-_44123734 | 0.29 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr12_+_53443963 | 0.29 |
ENST00000546602.1 ENST00000552570.1 ENST00000549700.1 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
| chr3_-_88108192 | 0.29 |
ENST00000309534.6 |
CGGBP1 |
CGG triplet repeat binding protein 1 |
| chr1_-_26232522 | 0.29 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr14_+_91580777 | 0.28 |
ENST00000525393.2 ENST00000428926.2 ENST00000517362.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr7_+_99699280 | 0.28 |
ENST00000421755.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr19_-_1568057 | 0.28 |
ENST00000402693.4 ENST00000388824.6 |
MEX3D |
mex-3 RNA binding family member D |
| chr8_-_38326119 | 0.28 |
ENST00000356207.5 ENST00000326324.6 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr20_-_39317868 | 0.28 |
ENST00000373313.2 |
MAFB |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr14_+_91580357 | 0.28 |
ENST00000298858.4 ENST00000521081.1 ENST00000520328.1 ENST00000256324.10 ENST00000524232.1 ENST00000522170.1 ENST00000519950.1 ENST00000523879.1 ENST00000521077.2 ENST00000518665.2 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr14_+_91580732 | 0.28 |
ENST00000519019.1 ENST00000523816.1 ENST00000517518.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr16_-_19533404 | 0.28 |
ENST00000353258.3 |
GDE1 |
glycerophosphodiester phosphodiesterase 1 |
| chr16_+_27561449 | 0.27 |
ENST00000261588.4 |
KIAA0556 |
KIAA0556 |
| chr1_-_113247543 | 0.27 |
ENST00000414971.1 ENST00000534717.1 |
RHOC |
ras homolog family member C |
| chr8_+_133787586 | 0.27 |
ENST00000395379.1 ENST00000395386.2 ENST00000337920.4 |
PHF20L1 |
PHD finger protein 20-like 1 |
| chr3_-_11623804 | 0.26 |
ENST00000451674.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr14_+_91580708 | 0.26 |
ENST00000518868.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr15_+_41136216 | 0.26 |
ENST00000562057.1 ENST00000344051.4 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr7_-_16685422 | 0.26 |
ENST00000306999.2 |
ANKMY2 |
ankyrin repeat and MYND domain containing 2 |
| chr19_+_36359341 | 0.26 |
ENST00000221891.4 |
APLP1 |
amyloid beta (A4) precursor-like protein 1 |
| chr7_+_100450328 | 0.25 |
ENST00000540482.1 ENST00000418037.1 ENST00000428758.1 ENST00000275729.3 ENST00000415287.1 ENST00000354161.3 ENST00000416675.1 |
SLC12A9 |
solute carrier family 12, member 9 |
| chr7_+_99699179 | 0.25 |
ENST00000438383.1 ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
| chr9_+_35538616 | 0.25 |
ENST00000455600.1 |
RUSC2 |
RUN and SH3 domain containing 2 |
| chr11_+_126225529 | 0.25 |
ENST00000227495.6 ENST00000444328.2 ENST00000356132.4 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr6_+_89790459 | 0.25 |
ENST00000369472.1 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
| chr2_+_220492116 | 0.25 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr11_+_64949899 | 0.25 |
ENST00000531068.1 ENST00000527699.1 ENST00000533909.1 ENST00000527323.1 |
CAPN1 |
calpain 1, (mu/I) large subunit |
| chr14_+_91581011 | 0.25 |
ENST00000523894.1 ENST00000522322.1 ENST00000523771.1 |
C14orf159 |
chromosome 14 open reading frame 159 |
| chr1_-_243418621 | 0.24 |
ENST00000366544.1 ENST00000366543.1 |
CEP170 |
centrosomal protein 170kDa |
| chr15_+_85144217 | 0.24 |
ENST00000540936.1 ENST00000448803.2 ENST00000546275.1 ENST00000546148.1 ENST00000442073.3 ENST00000334141.3 ENST00000358472.3 ENST00000502939.2 ENST00000379358.3 ENST00000327179.6 |
ZSCAN2 |
zinc finger and SCAN domain containing 2 |
| chr3_+_189507432 | 0.24 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr11_+_1860200 | 0.24 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr10_-_13390270 | 0.23 |
ENST00000378614.4 ENST00000545675.1 ENST00000327347.5 |
SEPHS1 |
selenophosphate synthetase 1 |
| chr9_+_34179003 | 0.23 |
ENST00000545103.1 ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1 |
ubiquitin associated protein 1 |
| chr2_+_220491973 | 0.23 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr16_-_53737795 | 0.22 |
ENST00000262135.4 ENST00000564374.1 ENST00000566096.1 |
RPGRIP1L |
RPGRIP1-like |
Gene Ontology Analysis
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.4 | 3.0 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.3 | 2.9 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.3 | 1.3 | GO:0047820 | D-glutamate cyclase activity(GO:0047820) |
| 0.3 | 0.8 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 3.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 1.4 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.2 | 5.1 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 0.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.2 | 1.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.2 | 4.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 3.6 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 0.6 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 0.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 2.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 3.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.1 | 1.0 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 1.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0004756 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.1 | 0.4 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.3 | GO:0004310 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.1 | 1.8 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.4 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.1 | 4.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.8 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 1.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.1 | 0.7 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.1 | 1.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.3 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.7 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0000995 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) transcription factor activity, core RNA polymerase III binding(GO:0000995) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 2.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.9 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.0 | 1.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 2.3 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.6 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 1.1 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.0 | GO:0004872 | receptor activity(GO:0004872) molecular transducer activity(GO:0060089) |
| 0.0 | 0.3 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 3.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.1 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 9.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 1.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 2.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 3.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 3.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.0 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.4 | 3.6 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.3 | 4.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.3 | 3.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.2 | 1.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 3.5 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 2.9 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.3 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.1 | 3.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 1.4 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.1 | 0.5 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.1 | 0.5 | GO:0001740 | Barr body(GO:0001740) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 3.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 1.4 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.1 | 2.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 1.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.5 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 4.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 3.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.3 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0071012 | catalytic step 1 spliceosome(GO:0071012) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 2.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.4 | GO:0097517 | stress fiber(GO:0001725) actin filament bundle(GO:0032432) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.9 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.5 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.4 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 2.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 4.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.9 | 3.6 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.5 | 1.4 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.4 | 1.8 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.4 | 1.2 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.4 | 3.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 4.5 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.4 | 2.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.4 | 2.9 | GO:0001845 | phagolysosome assembly(GO:0001845) negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.3 | 1.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 1.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.3 | 2.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.2 | 1.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.2 | 1.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.2 | 3.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.2 | 0.7 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.2 | 4.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 1.0 | GO:0030047 | actin modification(GO:0030047) |
| 0.2 | 1.0 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 | 2.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.2 | 0.5 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.2 | 1.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.2 | 0.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.5 | GO:2000830 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.1 | 3.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.4 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.1 | 1.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.5 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.9 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.1 | 0.3 | GO:0035283 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 0.3 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.1 | 0.8 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.8 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.2 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.1 | 0.8 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.1 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.7 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.3 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 1.0 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 1.0 | GO:0098903 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.6 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 | 0.7 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.7 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 2.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.7 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 2.0 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.3 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.1 | 0.3 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.1 | GO:2000586 | regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) |
| 0.1 | 0.2 | GO:1902075 | cellular response to salt(GO:1902075) response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.1 | GO:0021650 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve formation(GO:0021650) |
| 0.0 | 0.4 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) |
| 0.0 | 1.3 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 2.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.8 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 1.0 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.3 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 1.2 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.4 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.3 | GO:0040034 | regulation of development, heterochronic(GO:0040034) regulation of timing of cell differentiation(GO:0048505) |
| 0.0 | 0.9 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 1.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0048630 | skeletal muscle tissue growth(GO:0048630) |
| 0.0 | 0.1 | GO:0033023 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.3 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.2 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 0.4 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 1.0 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.0 | 0.1 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.6 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.3 | GO:0051683 | establishment of Golgi localization(GO:0051683) Golgi reassembly(GO:0090168) |
| 0.0 | 0.6 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 1.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.4 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.2 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.0 | 0.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0006536 | glutamate metabolic process(GO:0006536) |
| 0.0 | 0.5 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.4 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 1.4 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 0.1 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0030220 | platelet formation(GO:0030220) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 3.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 3.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 5.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 5.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 2.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.4 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 4.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 1.8 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.6 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |